3464
Patch2Self denoising of diffusion MRI in the cervical spinal cord improves repeatability and feature conspicuity1Radiology and Radiological Sciences, Vanderbilt University Medical Center, Nashville, TN, United States, 2Vanderbilt University Institute of Imaging Science, Vanderbilt University Medical Center, Nashville, TN, United States, 3Intelligent Systems Engineering, Indiana University Bloomington, Bloomington, IN, United States, 4Electrical Engineering and Computer Science, Vanderbilt University, Nashville, TN, United States
Synopsis
Diffusion MRI (dMRI) is a promising tool for evaluating the spinal cord in health and disease, however low SNR can impede accurate, repeatable, quantitative measurements. Here, we apply a recently proposed denoiser, Patch2Self, that strictly suppresses statistically independent random fluctuations in the signal originating from various sources of noise. Typical spinal cord dMRI scans have a smaller number of gradient directions (10-20) making PCA based 4D denoisers (require at least 30) inapplicable. Using self-supervised learning, Patch2Self addresses these issues which we quantitatively show with an improvement in repeatability and conspicuity of pathology in the spinal cord.
Introduction
Spinal cord (SC) diffusion MRI (dMRI) is challenging due to scan time, small size, motion and susceptibility artifacts resulting in lower SNR which impacts quantitative analysis. Denoising approaches have been applied to brain diffusion and improve SNR, reproducibility, precision, accuracy, and contrast. Commonly used single-subject denoisers such as PCA based methods1,2 assume low-rank, non-local methods3,4 leverage signal repetition, and the total variation norm5 presume the signal is locally smooth. PCA-based methods have been used most often on dMRI, but are not applicable to SC dMRI due to reduced gradient directions.Patch2Self6 does not assume the signal structure or the noise; only that the noise is random and uncorrelated across different gradient directions. Patch2Self holds out one volume and uses patches from all other volumes to predict the center of the patches of the held out volume using a J-invariant6 regressor. The noise is statistically independent, so the regressor learns the signal structure which exhibits some correlation and not the noise.
Here, we apply Patch2Self (implemented in DIPY7) to diffusion tensor imaging (DTI) in the cervical SC in healthy controls and multiple sclerosis (MS) patients. We show (1) improved goodness of fit of the diffusion signal, (2) improved repeatability of quantitative indices, and (3) increased lesion contrast.
Methods
Image AcquisitionThis study contains 3 datasets: 1) N=10 healthy controls (HC) (22-40yr, 5F/5M) with two sessions 3-5 months apart (inter-session repeatability); 2) one HC (34yr, F) with 5 repeated dMRI acquisitions within one session (intra-session repeatability); and 3) N=16 relapsing-remitting MS patients (20-42yr, 9F/7M, Expanded Disability Status Scale scores 0-1.5) with one session per patient.
Imaging was performed using a 3T Philips Elition MR scanner with 2-channel transmit and a dStream neurovascular coil (Philips) for reception. dMRI was acquired for 14 axial slices, cardiac-triggered, reduced field-of-view (FOV), single-shot EPI centered at C3/C4 level with FOV=80x57.5x70mm3, resolution=1.1x1.1x5mm3, SENSE (RL)=1.8, TR=5 beats (~4000ms), TE=77ms, NSA=3. A single-shell acquisition was used with 15 directions at b=750s/mm2. A high resolution (0.65x0.65x5mm3) anatomical, multi-echo, gradient echo (mFFE) image was acquired (TR/TE1/ΔTE=700/8.0/9.2ms) for the same 14 axial slices.
Processing
DTI volumes were motion-corrected using Spinal Cord Toolbox (SCT)8. Patch2Self6 was applied to cropped volumes. Quantitative DTI parameters (fractional anisotropy (FA), axial diffusivity (AD), radial diffusivity (RD), and mean diffusivity (MD)) were calculated using FSL for raw and denoised data. The DTI b0 image and parameter maps were registered to the mFFE and PAM50 template using SCT multimodal registration. Figure 1 summarizes the pre-processing steps.
Inter-session dataset: To evaluate effectiveness of Patch2Self denoising, we performed a voxel level cross-validation9 and computed goodness-of-fit for each voxel using the DTI model for the ten healthy controls with two scans.
Intra-session dataset: The Pearson correlation coefficient was computed for each possible pairing of the 5 intra-session dMRI acquisitions for the raw and denoised diffusion volumes. Mutual information (MI) was computed for FA for each pair of acquisitions to evaluate the effect of denoising on repeatability of a quantitative DTI metric.
MS patient dataset: Patch2Self was applied to dMRI data from MS patients to examine the effect of denoising on conspicuity of pathology (i.e. lesions) in diffusion volumes and quantitative DTI metrics.
Results
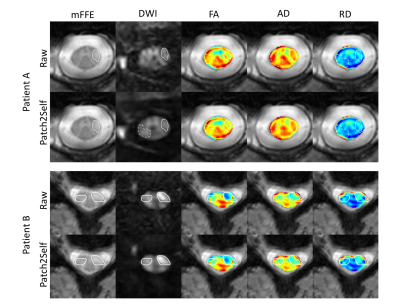
The goodness-of-fit for DTI model fitting improves significantly after Patch2Self denoising for 10 healthy subjects (R2 plots in Fig. 2). We see that due to noise suppression using Patch2Self, signal correlation between intra-session acquisitions improves significantly for each case along with reduced variation in the image intensities (Pearson correlation coefficient in Fig. 3). Additionally, the MI metric shows that FA becomes more repeatable between different acquisitions within the same session (Fig. 4).Denoising improves lesion conspicuity in diffusion images in MS patients as shown in two representative patients (Fig. 5). Of note, areas of suspected pathology have higher contrast after denoising, even when tissue appears “normal” in the corresponding mFFE anatomical image.
Discussion
Patch2Self incorporates non-local information into the same regression using self-supervised learning and works on a single subject. A unique advantage is that it can be applied at any pre-processing step only assuming that the noise is uncorrelated. We demonstrated that Patch2Self improves the signal modeling and downstream quantitative diffusion indices within the SC. Repeatability and conspicuity of pathology are key to deriving robust, quantitative dMRI biomarkers for diagnosis, predicting progression, and monitoring therapeutic response in diseases afflicting the SC such as MS. In the future, Patch2Self denoising of multi-shell dMRI data for advanced modeling approaches10,11 for the spinal cord can be explored to improve biological specificity to pathological processes.Acknowledgements
The authors thank all study participants, the VUIIS MRI technologists, and Dr. Anna Combes. This work was supported by the National Institutes of Health under award numbers 5R01NS109114 (S.A.S.), R01EY023240 (S.A.S.), R01EB017230 (B.A.L.), R01EB027585 (S.F. and E.G.), KL2TR002245 (K.P.O.), and K01EB030039 (K.P.O.), by the National Multiple Sclerosis Society award number RG-1501-02840 (S.A.S.), the Conrad Hilton Foundation (S.A.S.) and in part by the National Center for Research Resources grant UL1RR024975-0.References
[1] Veraart, Jelle, et al. "Denoising of diffusion MRI using random matrix theory." Neuroimage 142 (2016): 394-406.
[2] Ramos-Llordén, Gabriel, et al. "SNR-enhanced diffusion MRI with structure-preserving low-rank denoising in reproducing kernel Hilbert spaces." arXiv preprint arXiv:2009.06600 (2020).
[3] Coupé, Pierrick, et al. "An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images." IEEE transactions on medical imaging 27.4 (2008): 425-441.
[4] Dabov, Kostadin, et al. "BM3D image denoising with shape-adaptive principal component analysis." 2009.
[5] Knoll, Florian, et al. "Second order total generalized variation (TGV) for MRI." Magnetic resonance in medicine 65.2 (2011): 480-491.
[6] Fadnavis, Shreyas, Joshua Batson, and Eleftherios Garyfallidis. "Patch2Self: Denoising Diffusion MRI with Self-Supervised Learning." Advances in Neural Information Processing Systems 33 (2020).
[7] Garyfallidis, Eleftherios, et al. "Dipy, a library for the analysis of diffusion MRI data." Frontiers in neuroinformatics 8 (2014): 8.
[8] De Leener, Benjamin, et al. "SCT: Spinal Cord Toolbox, an open-source software for processing spinal cord MRI data." Neuroimage 145 (2017): 24-43.
[9] Rokem, Ariel, et al. "Evaluating the accuracy of diffusion MRI models in white matter." PloS one 10.4 (2015): e0123272.
[10] Lakhani, Dhairya A, et al. “Advanced multicompartment diffusion MRI models and their application in multiple sclerosis.” American Journal of Neuroradiology 41.5 (2020): 751-757.
[11] Schilling, Kurt G, et al. “Diffusion MRI microstructural models in the cervical spinal cord – Application, normative values, and correlations with histological analysis.” NeuroImage 201 (2019): 116026.
Figures