3434
Self-supervised IVIM DWI parameter estimation with a physics based forward model1Computational Radiology Laboratory, Boston Children's Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
Synopsis
The goal of this study was to assess the robustness and repeatability of intravoxel incoherent motion model (IVIM) parameter estimation for the diffusion weighted MRI in the abdominal organs under the constraints of noisy diffusion signal using a novel neural network training method. The method is based on the principle of a physics guided self-supervised neural network that does not require supervision for training. Such approach is beneficial in conditions where the reference methods are not available, or are not robust enough to provide good supervision. This work is targeting evaluations towards accelerated IVIM DWI scanning which exhibit low SNR.
Introduction
The abdominal diffusion weighted MR (DW-MR) images are inherently low in SNR reducing the robustness, and repeatability of the estimated parameters of the intravoxel incoherent motion model (IVIM) model [1,2]. However, increasing SNR requires increasing the repetitions acquired at each b-value which is clinically prohibitive, especially in pediatric imaging where increasing the scan time is not desirable. We introduce a deep learning algorithm aimed at improving the robustness and repeatability of IVIM parameter estimation for the DW-MRI in the abdominal organs under the constraints of noisy diffusion signal using a novel neural network. Due to lack of ground truth IVIM parameters for training a supervised network, we instead introduce a self-supervised 2D network.Methods
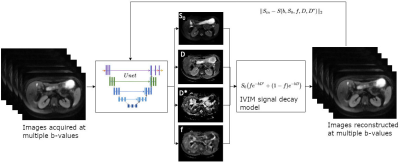
DW-MR images from 84 pediatric Crohn's disease patients were acquired clinically at 1.5T using a free-breathing single-shot EPI: TR/TE=7500/77ms; matrix=192x156; FOV=300x260mm; slice=5mm; b-values=0,50,100,200,400,600,800 s/mm^2 with 6 diffusion gradient directions and 1 repetition; acquisition time~= 5.5min. In these images, we analyzed the regions segmented in kidney cortex, spleen and liver. A novel training regime was designed for a U-net style neural network [Fig 1]. The inputs to the Unet consisted of 7 b-value images that were obtained by geometrically averaging 6 directional diffusion images. The outputs of the network were the 4 parameter map images of the IVIM - pseudo-diffusion coefficient (D), fast diffusion coefficient (D*), pseudo-diffusion fraction (f) and S0. The network predicted parameter maps were propagated through the IVIM forward model, as tensors in the tensorflow graph, and an estimate of the input signal was computed. This estimate was compared to the measured input signal with a L2-norm loss, hence forming a self supervised training loop. No reference labels, such as estimates from a conventional scan was necessitated for this approach, as is typically done for supervised training. The data was split into 60, 4, 20 scans for training, validation and test respectively. Hyperparameters were: dropout=0.1; batch=2; Adam optimizer; learning rate of 0.0008 with progressive decreasing schedule; epochs=80. The network was designed in Tensorflow 1.3.0 and trained on Nvidia TitanXP GPU. Inputs were preprocessed by threshold based masking of the body in axial DW-MRI and normalization in the 0-1 scale. 20 test scans were noise corrupted to evaluate performance at reduced SNR conditions. A dataset from 5 healthy volunteers with repeated intrasession acquisitions was evaluated for assessment of reproducibility. The method was compared to: a conventional non-linear least squares (NLLS) based IVIM fitting method with segmented fitting [3] and bounded constraints based on the BOBYQA method (IVIM) [4]; a synthetically trained voxelwise multi-layer perceptron network (DeepIVIM) [5].Results
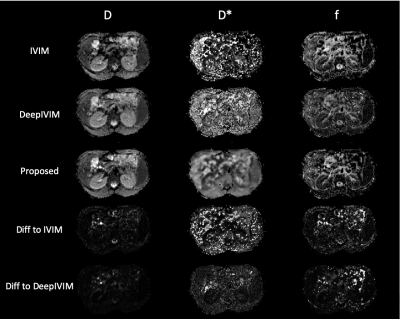
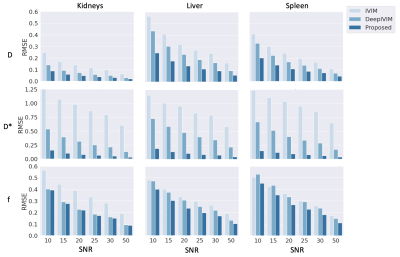
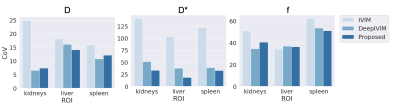
Qualitative assessment of the estimated parameter maps show agreement with NLLS and DeepIVIM methods in D and f, while the estimates for D* are significantly more smooth [Fig 2]. The proposed method yields a large reduction in variability of D* estimates under non-noisy input signal when compared to standard IVIM [Fig 3]. Statistical analysis with the Mann-Whitney-Wilcoxon non-parametric test shows no significant difference between the proposed method and IVIM for non-noisy input for D. Significant difference is observed in D* of liver and f of kidneys, which is also attributed in the comparison between DeepIVIM to IVIM. The proposed method increased the robustness of parameter estimations in the presence of increased noise when evaluated against IVIM and DeepIVIM [Fig 4]. Normalized RMSE measures, calculated over all individual available voxels, are lowered for D* by at least half in the proposed method when compared to DeepIVIM, and are lowered by almost 10x when compared to IVIM. A significant reduction in nRMSE across different noise levels is also achieved for the estimates of f and D, although it is less pronounced. In the repeatability experiment, the proposed method shows reduced coefficient of variation (CoV) of the fast diffusion rate estimates over multiple acquisition in comparison to conventional IVIM (from 140\% to 30\% in kidneys and from 100\% to 20\% in liver) and comparable performance to DeepIVIM [Fig 5]. A reduced CoV is achieved also for kidneys and spleen, but not for liver for f, where the CoV measures between all three methods are in close proximity.Discussion
The method achieves significant gain in robustness of the D* parameter estimates that is particularly difficult to estimate with the conventional methods. Results indicate robust performance under the conditions of gaussian normal noise for all parameters. The study was designed with scan acceleration in mind, where IVIM data would be acquired with a smaller number of directional diffusion gradients or repetitions, thereby causing a reduction in SNR but also results in a faster scan. As evidenced by the experiments, the proposed method was more resistant to lower SNRs than the conventional approach and voxelwise neural network. It should be noted that when we reduce number of measurements averaged at each b-value, the average measurements become more sensitive to physiological motion. Motion between images acquired at different b-values and directional gradients remains to be an active issue that needs to be addressed.Conclusion
The proposed method yields robust performance under the constraints of noisy DW-MR images. The increased accuracy of parameter estimates indicates a potential for use in accelerated abdominal DW-MRI acquisition which will exhibit even lower SNR.Acknowledgements
This work was supported partially by the Boston Children's Hospital Translational Research Program Pilot Grant 2018, Society of Pediatric Radiology Multi-center Research Grant 2019, Crohn’s and Colitis Foundation of America’s (CCFA) Career Development Award and by the National Institutes of Health under award numbers R01EB019483, R21DK123569, R21EB029627, and by the grant No 2019056 from the United States-Israel Binational Science Foundation (BSF), and a pilot grant from National Multiple Sclerosis Society under Award Number PP-1905-34002.References
1) Le Bihan D, Breton E, Lallemand D, Aubin M, Vignaud J, Laval-Jeantet M. 1988. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 168:497–505
2) Koh DM, Collins DJ. 2007. Diffusion-weighted MRI in the body: applications and challenges in oncology. American Journal of Roentgenology 188:1622–1635
3) Marquardt DW. 1963. An algorithm for least-squares estimation of nonlinear parameters. Journal of the society for Industrial and Applied Mathematics 11:431–441.
4) Powell MJ. 2009. The BOBYQA algorithm for bound constrained optimization without deriva-350tives. Cambridge NA Report NA2009/06, University of Cambridge, Cambridge :26–46
5) Barbieri S, Gurney-Champion OJ, Klaassen R, Thoeny HC. 2020. Deep learning how to fit an intravoxel incoherent motion model to diffusion-weighted MRI. Magnetic resonance in medicine;83:312–321
Figures