3427
Deep Learner estimated isotropic volume fraction enables reliable single-shell NODDI reconstruction1Department of Electrical and Computer Engineering, University of Rochester, Rochester, NY, United States, 2Department of Imaging Sciences, University of Rochester, Rochester, NY, United States, 3Department of Biomedical Engineering, University of Rochester, Rochester, NY, United States, 4Department of Neurology, University of Rochester, Rochester, NY, United States, 5Department of Physics and Astronomy, University of Rochester, Rochester, NY, United States
Synopsis
The study explores the possibility to create neurite orientation dispersion and density imaging (NODDI) parameter maps with single-shell diffusion MRI data, using isotropic volume fraction (fISO) as prior. Proposed dictionary based deep learning prior NODDI (DLpN) framework leverages fISO estimation with a diffusion imaging scalar and T2 weighted non-diffusion signal. fISO estimation was validated in simulation and in-vivo. DLpN derived NDI and ODI maps for single-shell protocols are comparable with original multi-shell NODDI (error <5%) and may allow NODDI evaluation of retrospective brain studies on single-shell diffusion MRI data by multi-shell scanning of two subjects for DictNet fISO training.
Introduction
Neurite orientation dispersion and density imaging (NODDI) is increasingly popular in understanding brain microstructures with multi-shell diffusion MRI (dMRI)1. However, single-shell NODDI reconstruction has been held back in literature as it failed to fit relevant parameters, especially neurite density index (NDI)1. Previous works showed to map diffusion tensor with NODDI parameters but no ground validation or margin of error was reported for isotropic volume fraction (fISO)>0 2,3. In this study, using both synthetic and in vivo data, we investigated if fISO estimation can be relieved from NODDI model constraint and used as a prior to enable single-shell NDI and orientation dispersion index (ODI) estimation similar to multi-shell protocol. First, we proposed stochastic sparse dictionary based deep learner (DictNet) to estimate reliable fISO validated with simulation. Then we compared the performance of single-shell reconstruction to multi-shell generated maps using our Deep Learning prior NODDI (DLpN) and original NODDI approach.Method & Materials
DictNet:Proposed DictNet is a stochastic sparse dictionary-based learning strategy that is devised from previous works4,5 with modified Iterative Hard Thresholding (IHT)6 approach to estimate fISO, utilizing FA and S0 in the dictionary to make the single-shell fISO estimation stable. Briefly, we train sparse dictionary $$$\Phi$$$, with following regularized problem,$$\hat{f}=\arg\min_{f \geq 0} || \Phi f^{S0}-y||^2_2 +\lambda' ||f||_0 $$
where, $$$\lambda'$$$ tunes sparsity level of $$$f$$$. In the following equation, Dictionary layers (L and D) utilize the first layer of Iterative Hard Thresholding (IHT) algorithm6 with stochastic vector initialization, d. Shown as follows- $$f=H_\lambda(L d_{in}+D d) $$
where, din is the dMRI input. For single-shell fitting, generated coefficient vector $$$f$$$ contributes with additional IHT layer for FA and, T2w or S0 layers and contribute in the final estimation through thresholder $$$H_{\lambda}(\cdot)$$$.
$$f^{S0}=H_{\lambda}(FA\cdot L_{FA} + S0 \cdot L_{S0} + f\cdot L_{f})$$
where, $$$L_{FA},L_{S0}, L_{f} $$$ weighs FA, S0 and $$$f$$$ respectively in single shell fISO learning. Multi-shell learning with DictNet only utilizes $$$f$$$ in the learning process. $$$f$$$ or $$$f^{S0}$$$ finally maps fISO with a feedforward network.
DLpN Framework: Generating NDI and ODI maps
The Rician loglikelihood framework from NODDI was modified for DLpN to account for prior fISO, thereby used to generate NDI and ODI maps.
In-vivo and Simulated Synthetic Data:
We have used de-identified dMRI data from 8 randomly selected subjects from the Human Connectome Project (HCP) dataset7. Images were collected at 3T (Connectome, Siemens, Erlangen, Germany) using SE-EPI sequence for 1.25mm isotropic resolution; and 90 gradient directions for each shell [b-values=1000 (P1), 2000 (P2), and 3000 (P3) s/mm2], and 18 b=0 s/mm2 reference images. Details of the protocol and preprocessing can be found in 7,8. Multi-shell protocol [i.e., b=1000,2000,3000 s/mm2 (Pall)] was used to generate the ground-truth NODDI parameters in vivo using the NODDI toolbox (v1.01)1.
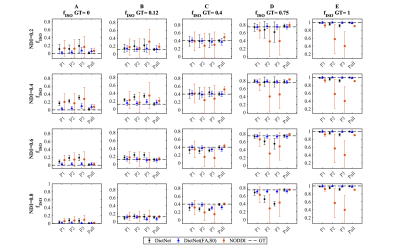
Using NODDI equation1 similar to original NODDI model, dMRI data was synthesized with following parameters (NDI=[0.2, 0.4, 0.6, 0.8]; fISO=[ 0, 0.12, 0.4, 0.75, 1]; Axon radii=[ 0.5, 1, 2, 4] μm; k=[0, 0.25, 1, 4,16], with 254 different orientations) with 20dB Rician noise to report the stability of fISO reconstruction with DictNet with and, without FA and S0 side by side with NODDI for protocols P1, P2, P3 and Pall.
DictNet- Training, Cross-Validation and Test:
For training DictNet, we used the ground-truth fISO from simulation, and multi-shell NODDI generated fISO from 2 HCP datasets; 15% of the voxel-wise training data were used for cross-validation. The tests were performed on a synthetic test set and the 6 remaining HCP subjects in respective cases. All the Harvard-Oxford (grey matter) and JHU-ICBM (white matter) structures were transformed to subject space using ANTs9 and used to report region-wise error calculation on test data set.
Results and Discussion
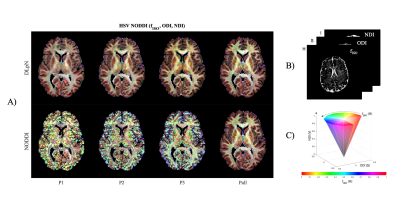
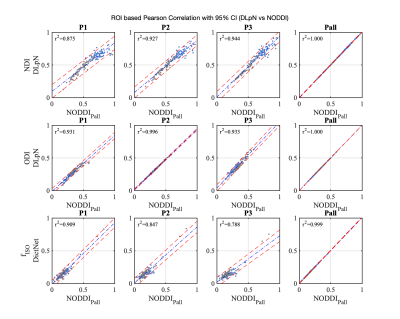
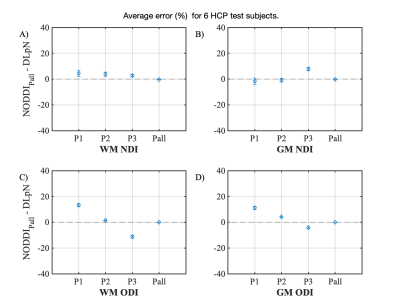
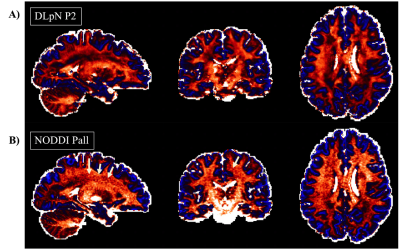
Figure 1 compares the simulation results for fISO estimation with DictNet variants and NODDI. DictNet generated fISO revealed to be reliable when FA and S0 is utilized to build the dictionary. Figure 2 presents the in vivo results in HSV (Hue, Saturation, Value) and it is evident that DLpN performs better in comparison with NODDI for single-shell and analogous with multi-shell; also, the DLpN single-shell estimated results are comparable with multi-shell NODDIPall. The correlation with pseudo ground truth NODDIPall, shows r2>0.9 for NDI and ODI and, r2>0.84 for fISO (Figure 3). Figure 4 reports NDI and ODI mean percentage error for defined protocols with DLpN reconstruction for 6 HCP test data compared to NODDIPall. Our results suggest that DLpN estimated NDI and ODI parameters for single-shell protocols are comparable with original multi-shell NODDI, and protocol with b=2000 s/mm2 performs the best, where the reported error with respect to multi-shell NODDIPall was ~2% in white matter and ~4% in grey matter. Figure 5 illustrates a close comparison between DLpNP2 and NODDIPall NDI, highlighting plausible noisy overestimation with multi-shell NODDI.Conclusion
The DLpN single-shell derived NDI and ODI maps are analogous with those reconstructed with the original multi-shell NODDI model, especially with b=2000 s/mm2. Since fISO prior estimation is shown to be independent and well-posed when deep learning dictionary is constructed using corresponding FA and S0, this approach may allow conventional studies on single-shell dMRI data to evaluate NDI and ODI with the prerequisite of two additional scans for training DictNet.Acknowledgements
This work was supported by the National Institutes of Health (Grant Numbers: R01AG054328, and R01MH118020).References
1. Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61(4):1000-1016.
2. Alexander DC, Zikic D, Ghosh A, Tanno R, Wottschel V, Zhang J, et al. Image quality transfer and applications in diffusion MRI. Neuroimage 2017; 152:283-298.
3. Edwards LJ, Pine KJ, Ellerbrock I, Weiskopf N, Mohammadi S. NODDI-DTI: estimating neurite orientation and dispersion parameters from a diffusion tensor in healthy white matter. Frontiers in neuroscience 2017; 11:720.
4. Ye C. Estimation of tissue microstructure using a deep network inspired by a sparse reconstruction framework. In: International Conference on Information Processing in Medical Imaging: Springer; 2017. pp. 466-477.
5. Ye C, Li X, Chen J. A deep network for tissue microstructure estimation using modified LSTM units. Medical image analysis 2019; 55:49-64.
6. Blumensath T, Davies ME. Iterative Thresholding for Sparse Approximations. Journal of Fourier Analysis and Applications 2008; 14(5):629-654.
7. Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: An overview. NeuroImage 2013; 80:62-79.
8. Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens T, Bucholz R, et al. The Human Connectome Project: a data acquisition perspective. Neuroimage 2012; 62(4):2222-2231.
9. Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011; 54(3):2033-2044.
Figures