3405
DIFFnet: Diffusion parameter mapping network generalized for input diffusion gradient directions and b-values1Seoul National University, Seoul, Korea, Republic of, 2Hankuk University of Foreign Studies, Gyeonggi-do, Korea, Republic of
Synopsis
A deep neural network, referred to as DIFFnet, was developed to reconstruct the diffusion parameters from data with reasonable b-value and gradient scheme (gradient direction and the number of gradients). For the generalization, Qmatrix was proposed via the projection and quantization of q-space. DIFFnet was trained by simulated datasets with various b-values and gradient schemes. Two DIFFnets, one for DTI and the other for NODDI were developed. DIFFnet successfully reconstructs the diffusion parameter maps of two in-vivo datasets with different b-values and gradient schemes.
Introduction
Recently, several deep neural networks have been presented for diffusion parameter mapping1-3. However, these networks allow us to input fixed b-values and fixed diffusion gradient schemes (i.e., gradient direction and the number of gradients) that were used for training. Hence, if data with different b-values or gradient schemes exist, the networks need to be retrained with a new training dataset, requiring long preparation time and efforts. This issue of generalization for the input data is critical for neural networks. In this work, we present a new deep neural network, which is referred to as DIFFnet, that allows us to reconstruct diffusion model parameters from various b-values and gradient schemes. Two DIFFnets were designed: DIFFnetDTI for diffusion tensor imaging (DTI)4,5 and DIFFnetNODDI for neurite orientation dispersion and density imaging (NODDI)6. In each model, two in-vivo datasets, which have different b-values and gradient schemes, are evaluated for performance.Method
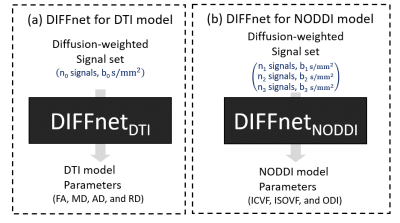
[DIFFnet outline] For DTI (Fig. 1a), DIFFnetDTI was constructed to generate DTI parameters from a diffusion signal set with $$$n_0$$$ number of diffusion gradient directions with a single b-value of $$$b_0$$$ s/mm2. For NODDI (Fig. 1b), DIFFnetNODDI was designed to generate NODDI model parameters from a three-shell protocol, such as $$$n_1$$$, $$$n_2$$$, and $$$n_3$$$ diffusion-weighted signals for b-value of $$$b_1$$$, $$$b_2$$$, and $$$b_3$$$ respectively. DIFFnet was aimed to generate the parameter maps from various b-values, diffusion gradient direction, and the number of gradients.To achieve this goal, a Qmatrix was introduced and utilized for an input of DIFFnet: First, the normalized signals were placed in a q-space with a q-vector as spatial coordinates (Fig. 2b). For DTI, signals were projected on a xy-, yz-, and xz-plane (Fig. 2c), quantized along the two axes by $$$q_n$$$ and concatenated, producing $$$q_n × q_n × 3$$$ matrix. Five different $$$q_n$$$ values, 5, 10, 15, 20, and 25, were tested. For NODDI (Fig. 2c), the projection was performed on each shell, quantized by $$$q_n$$$, and concatenated, producing $$$q_n × q_n × 9$$$ matrix. As a structure of DIFFnet, ResNet was utilized7.[Training dataset generation] Monte-Carlo diffusion simulation was performed to generate the training dataset. For DIFFnetDTI, to obtain signal set with various gradient schemes, $$$n_0$$$ and $$$b_0$$$ were randomly selected in the range of 30 to 80 and 600 to 1200 s/mm2, respectively. For DIFFnetNODDI $$$n_1$$$, $$$n_2$$$, and $$$n_3$$$ were chosen in the ranges of 5 to 10, 25 to 50, and 50 to 100, respectively. Those for $$$b_1$$$, $$$b_2$$$, and $$$b_3$$$ were sampled in the ranges of 250 to 350, 600 to 800, and 1800 to 2200 s/mm2, respectively. A total of 106 protons, assumed to have an unit magnetization each, were generated and performed random walks, following DTI or NODDI diffusion model. The phase of magnetization for each spin was accumulated. After the simulation was performed, a complex-averaged signal was calculated based on the pulsed gradient spin echo sequence. A total of 106 times of simulations were conducted for training in both DTI and NODDI.
[Dataset acquisition & processing] A total of four datasets, with five in-vivo scans each, were used for evaluation. For DTI, DatasetDTI-A of b = 700 s/mm2 with 32 directions and DatasetDTI-B of b = 1000 s/mm2 with 30 directions were utilized. For reference, conventioanl DTI maps were reconstructed4. For NODDI, DatasetNODDI-A of b = 300, 700, and 2000 s/mm2 with 8, 32, and 64 directions, and DatasetNODDI-B of b = 300, 700, and 2000 s/mm2 with 8, 30, and 60 directions were used. For reference, conventional NODDI maps were reconstructed6.
[Evaluation] In DTI, DIFFnetDTI was evaluated with DatasetDTI-A and DatasetDTI-B. For the comparison, a multi-layer perceptron (MLP)2 trained by a simulated dataset having a gradient scheme of DatasetDTI-A was compared. Similarly, DIFFnetNODDI and MLP, trained by a dataset having a gradient scheme of DatasetNODDI-A, was tested with DatasetNODDI-A and DatasetNODDI-B. Additionally, accelerated microstructure imaging via convex optimization (AMICO) was tested.
Results
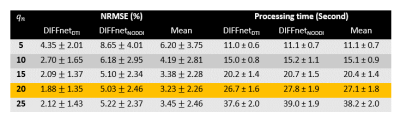
When the effects of five $$$q_n$$$ is investigated (Fig. 3), the results reveal that $$$q_n$$$ of 20 shows the minimum NRMSE, and, therefore, $$$q_n$$$ of 20 is chosen as the default for DIFFnet. In the reconstruction of the DTI maps, DIFFnetDTI generates highly accurate DTI maps in both datasets (Fig. 4). On the other hand, MLP fails to reconstruct DatasetDTI-B, which has a different gradient scheme from the training dataset of MLP, demonstrating the importance of the input type generalization in neural networks. Similar trends can be observed in NODDI reconstruction. DIFFnetNODDI successfully generates the NODDI maps of the two datasets with different gradient schemes (Fig. 5), whereas MLP fails to reconstruct DatasetNODDI-B. Compared to AMICO results (mean NRMSEs of ICVF:6.24±0.51%, ISOVF:7.21±0.93%, and ODI:8.82±1.25%), DIFFnetNODDI show lower mean NRMSEs (ICVF:3.78±0.37%, ISOVF:3.61±0.47%, and ODI:7.89±0.44%). Processing time of DIFFnet is measured to be less than 30seconds for all experiments.Discussion and Conclusion
In this study, DIFFnet was developed to reconstruct the diffusion model parameters from data with various b-value and gradient scheme. Different from previously proposed deep learning methods, which utilized fixed b-values and fixed diffusion gradient schemes, DIFFnet does not specify gradient directions and b-values for its input dataset. In the experiments of DTI and NODDI, DIFFnet demonstrated successful reconstruction for different b-values and gradient scheme datasets.Acknowledgements
This research was supported by the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (NRF-2017M3C7A1047864) and NRF grant funded by the Korea government (MSIT) (NRF-2018R1A4A1025891).References
[1] Y. Masutani, "Noise Level Matching Improves Robustness of Diffusion Mri Parameter Inference by Synthetic Q-Space Learning," in 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), 2019: IEEE, pp. 139-142.
[2] V. Golkov et al., "q-Space Deep Learning: Twelve-Fold Shorter and Model-Free Diffusion MRI Scans," IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1344-1351, 2016, doi: 10.1109/TMI.2016.2551324.
[3] C. Ye, Y. Cui, and X. Li, "Q-space learning with synthesized training data," in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2019: Springer, pp. 123-132.
[4] P. J. Basser, J. Mattiello, and D. LeBihan, "MR diffusion tensor spectroscopy and imaging," (in eng), Biophys J, vol. 66, no. 1, pp. 259-67, Jan 1994, doi: 10.1016/s0006-3495(94)80775-1.
[5] P. J. Basser, J. Mattiello, and D. Lebihan, "Estimation of the Effective Self-Diffusion Tensor from the NMR Spin Echo," Journal of Magnetic Resonance, Series B, vol. 103, no. 3, pp. 247-254, 1994/03/01/ 1994, doi: https://doi.org/10.1006/jmrb.1994.1037.
[6] H. Zhang, T. Schneider, C. A. Wheeler-Kingshott, and D. C. Alexander, "NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain," (in eng), Neuroimage, vol. 61, no. 4, pp. 1000-16, Jul 16 2012, doi: 10.1016/j.neuroimage.2012.03.072.
[7] K. He, X. Zhang, S. Ren, and J. Sun, "Deep residual learning for image recognition," in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770-778.
Figures