3381
Shot-wise separate motion correction and ICA denoising for BISEPI high-resolution fMRI study at 7T1CiNet, NICT, Osaka, Japan, 2Graduate School of Frontier Biosciences, Osaka University, Osaka, Japan, 3Tohoku Fukushi University, Sendai, Japan
Synopsis
Shot-wise separate motion correction is presented for BISEPI (Block-Interleaved Segmented EPI)-based high-resolution fMRI at 7T. Artifacts caused by subject motion during longer acquisition time of one volume than normal multi-shot EPI reduces the performance of BISEPI. The proposed k-space motion correction (KMoCo) method calculates and corrects the motion during the shot-wise acquisition of each volume separately. We also performed ICA denoising on the KMoCo data. The proposed method improves temporal SNR, the number of detected active voxels, and localization of brain activity in high-resolution fMRI at 7T
Introduction
BISEPI (Block-Interleaved Segmented EPI) has been reported for high-resolution fMRI studies at 7T1. Artifacts caused by subject motion during longer acquisition time of one volume than normal multi-shot EPI reduces the performance of BISEPI. The proposed k-space motion correction (KMoCo) method calculates and corrects the motion during the shot-wise acquisition of each volume separately. Moreover, we investigated ICA denoising2 for the removal of motion-related artifacts after KMoCo in BISEPI acquired data.Methods
Two adult human brains were scanned using the BISEPI1 sequence in two different experiments on a Siemens MAGNETOM 7T scanner with a 32-channel phased-array head coil with TR = 1.5 s. One normal multi-shot EPI volume was acquired as a reference before BISEPI acquisition in block-design stimulation. In post-processing stage, each shot of BISEPI acquired data was reconstructed to images using GRAPPA3 (like single shot EPI), the motion information was then obtained using SPM-12 by comparing the difference between the reference image and each single shot image. Before the final reconstruction, BISEPI data was motion corrected shot-wise separately in k-space as illustrated in Fig. 1. After KMoCo, we performed ICA2 data exploration using Melodic ICA implemented in FSL 5.0.7. No additional spatial or temporal smoothing was performed. The data were subsequently denoised by regressing out the ICA derived noise components. BrainVoyager, FSL and Matlab were used for data pre-processing and analysis. In experiment 1, we acquired 320 volumes of 0.71 x 0.71 x 1.00-mm resolution fMRI data during a visual task of 30 s stimuli followed by 30 s rest, each repeated 8 times, with GRAPPA = 2, number of shot = 3. Each shot in BISEPI-acquired data-set was reconstructed to shot-separated volumes (GRAPPA = 6) for motion detection. In experiment 2, we acquired 300 volumes of 0.70 x 0.70 x 0.70-mm resolution data with number of shot = 3 and no GRAPPA during visual tasks of varying inter-stimulus intervals4 (ISIs) of 6 s stimuli followed by 24 s rest, each repeated 5 times.Results and Discussion
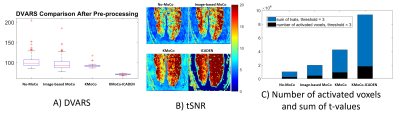
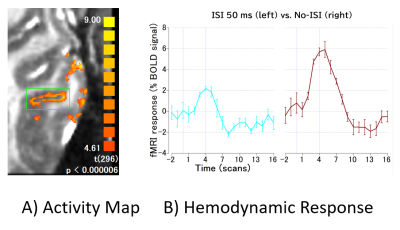
We compared the performance of the proposed method with no motion correction and conventional image-based motion correction. Fig. 2 (A) shows the comparison of the results in terms of the RMS variance over voxels of the temporal derivative of the time-series (DVARS)5 among image-based MoCo, the proposed KMoCo, and ICA denoising (ICADEN) for the data acquired in experiment 1. Fig. 2 (B-C) shows the temporal SNR (tSNR) gain and the number of detected active voxels as well as sum of t-values. Fig. 3 (A) shows the detected activity after performing KMoCo and ICADEN. Fig. 3 (B) shows the stimuli-specific hemodynamic response in the region indicated by the green rectangle on the activation map. The proposed approach substantially decreased motion outliers and improved the localization of brain activity thus providing benefit for the analysis of high-resolution fMRI data acquired using BISEPI at 7T.Acknowledgements
No acknowledgement found.References
1. Liu G, Shah A, Ueguchi T. Block-Interleaved Segmented EPI for voxel-wise high-resolution fMRI studies at 7T. Proc. Intl. Soc. Mag. Reson. Med. 26; 2018; 5450.
2. Beckmann CF and Smith SM. Probabilistic Independent Component Analysis for Functional Magnetic Resonance Imaging. IEEE Trans. on Medical Imaging; 2004; 23:2(137-152).
3. Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn. Reson. Med. 47(6); 2002; 1202–1210.
4. Sung YW, Kamba M, Ogawa S. Progression of neuronal processing of visual objects. Neuroreport. 2007 Mar 26;18(5):411-4.
5. Power JD, Barnes KA, Snyder AZ, et al. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. NeuroImage 59; 2012; 2142– 2154.
Figures
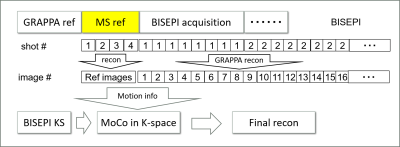
Figure 1: Procedure of the proposed k-space motion correction (KMoCo) in BISEPI acquired data.