3370
Where is the Pain in the Brain? Functional MRI of Saliency versus Nociception at 7.0 Tesla en route to Diagnostic Biomarkers of Pain1Berlin Ultrahigh Field Facility (B.U.F.F.), Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany, 2Spinoza Centre for Neuroimaging, Amsterdam, Netherlands, 3Experimental and Clinical Research Center (ECRC), a joint cooperation between the Charité Medical Faculty and the Max Delbrück Center for Molecular Medicine in the Helmholtz Association, Berlin, Germany
Synopsis
Identifying diagnostic biomarkers of pain is of utmost clinical importance. This is challenging since similar fMRI patterns are elicited by painful and equisalient non-painful stimuli at 3T. To decipher pain-specificity this work uses 7T high-resolution fMRI of (1) salient non-painful versus painful stimuli, (2) different types of painful stimuli in "healthy" controls versus (3) a subject with a genetically-induced pain-free phenotype. We found that canonical HRF modeling is insufficient to reliably capture the shape of elicited BOLD responses that further varied in correlation to perceived saliency.
Introduction
Pain is a major clinical, social and economical problem which constitutes the unmet need of efficacious strategies for diagnosis and treatment of this invisible burden: We do not know whether persons who cannot communicate, like coma patients or elderly people with Alzheimer’s are in pain or not, and chronic pain is one of the largest and most cost intensive diseases in the developed world1,2. Identifying fMRI biomarkers for pain is key to objective diagnosis and individualized treatment1,2. However, decoding pain-specific neurosignatures remains challenging due to the spatial resolution constraints at clinical field strengths. The so-called pain matrix is not specific to pain, but responds to all sorts of attention-grabbing stimuli2-4 – including nociceptive stimuli in subjects who cannot feel pain due to a genetic anomaly5. Recognizing this challenge the goal of this work is to disentangle saliency from pain by (1) comparing differences in patterns evoked by salient non-painful and painful stimuli at 7T using twice the spatial resolution commonly used at 3T. To further stress pain specificity in the detected patterns, we (2) compared different types of painful stimuli and (3) also included a subject who does not feel any pain due to a genetic anomaly. With this strategy we aim to identify pain-specific functional neurosignatures that may serve as a first step en route to diagnostic biomarkers of pain.Methods
Experimental design: 8 male volunteers (7 controls, 1 analgesic subject) underwent fMRI, consistent of 4 successive runs of 12 min each. Three different types of stimuli (8 per type and run; interstimulus spacing 18-24s) were applied in a pseudorandomized manner: Electrical stimulation (100 ms) was applied to the right ankle, over the superficial peroneal nerve in two different stimulus strengths: 6mA and 10 mA. Short pulses of transient heat (51°C) were applied to the dorsum of the right foot (CHEPS, Medoc Ltd.). MRI: High-resolution T2*-weighted fMRI (GE-EPI, TR/TE/FA = 2000ms/33.2ms/66°, FOV/matrix = 240x240mm/160x160, number of slices = 80, spatial resolution = 1.5mm isotropic, 220 volumes) was acquired on a MAGNETOM 7T scanner (Siemens Healthcare, Erlangen, Germany) with a single-channel-transmit/32-channel receive head coil (Nova Medical, Wilmington, MA, USA). Data analysis: fMRI data were motion and distortion corrected, smoothed, registered to the MNI152 template, and statistically analyzed using FSL FEAT. HRF were modeled for the individual stimuli, double gamma convolution + temporal derivative. Corrected cluster significance thresholds were determined by z > 3.1 (p<0.05).Results
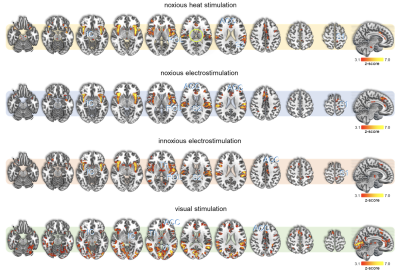
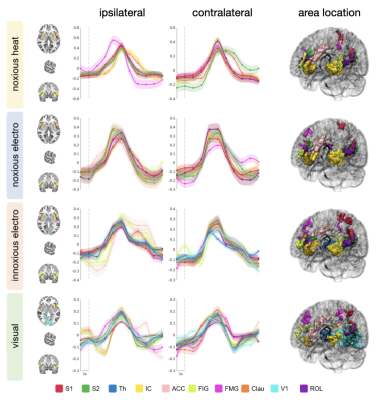
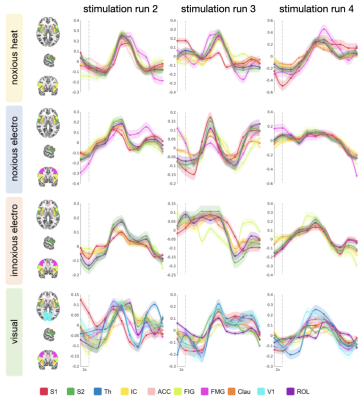
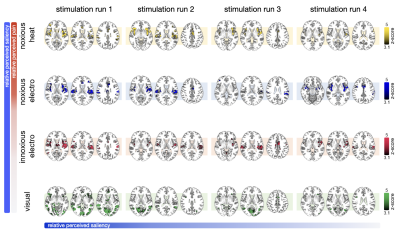
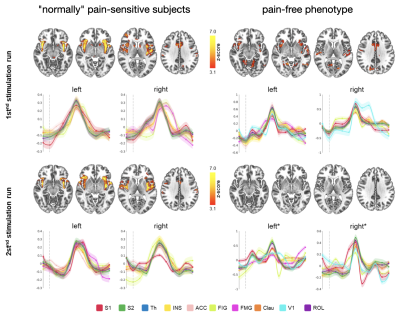
All stimuli were reported as salient, while the 10mA electrical and heat stimuli were reported as painful among "normally" pain-sensitive subjects. Their group analysis revealed a number of overlapping significant clusters across non-painful and painful stimuli in areas that comprise the classical pain matrix, including the secondary somatosensory cortex (S2), the anterior cingulate cortex (ACC), and the insula (Fig. 1). Differences to the former study at 3T include smaller clusters in comparison3, but this is partially due to different standards in thresholding6. We found less activity in the insula for visual stimuli, and no activity at all in the thalamus. The latter was surprising as the thalamus comprises the relay nuclei of the received sensory inputs. The BOLD responses occurred bilaterally. In the first run, hemodynamic response functions (HRF) were shaped quite similar across stimuli in both hemispheres (Fig. 2). With subsequent runs, the HRF got increasingly out of shape (Fig.3). This led to difficulties in modeling the HRF at later runs, which was reflected in the spatial maps (Fig. 4). Finally we did a case study in a subject who doesn’t feel neither temperature nor pain due to a genetic anomaly in 2 separate sessions using a study design and analysis similar to the one reported above (Fig. 5). Based on the subject’s report the heat stimulus was perceived as salient non-painful sensation. HRF to noxious heat were generally more variable in the pain-free subject, and were particularly badly captured by the canonical model in the subsequent runs. This lead to the absence of significant clusters for the applied statistic threshold, although we clearly see HRF responses to the noxious stimulus (Fig. 5).Summary and Conclusion
By employing a multi-modal event-related stimulation design with 7T high-resolution fMRI, we found canonical HRF modeling to be inapt to accurately capture BOLD responses to brief painful and non-painful salient stimuli across multiple runs. The HRF got out of shape with subsequent runs, which correlated to a loss of perceived saliency over time for all stimuli. This emphasizes a strong dependence on saliency, but also reveals a notable feature of multi-run paradigms for training machine learning classifiers to decode pain-specific neurosignatures4. The fact that we did not find any activity in the thalamus is probably due to a greater variability of HRF with smaller voxel size at 7T7. This warrants further research including the application of finite impulse responses, and multi-voxel pattern analysis.Acknowledgements
No acknowledgement found.References
1. Miesen M, Lindquist MA, Wager TD (2019). Neuroimaging-based biomarkers for pain: state of the field and current directions. PAIN Reports 4(4):e751 doi: 10.1097/PR9.0000000000000751.
2. Mouraux A, Iannetti D (2018). The search for pain biomarkers in the human brain. Brain 141(12). doi: 10.1093/brain/awy281.
3. Mouraux A, Diukova A, Lee MC, Wise RG, Iannetti GD (2011). A multisensory investigation of the functional significance of the "pain matrix“. Neuroimage 54(3):2237-49. doi: 10.1016/j.neuroimage.2010.09.084.
4. Liang M, Su Q, Mouraux A, Iannetti GD (2019). Spatial Patterns of Brain Activity Preferentially Reflecting Transient Pain and Stimulus Intensity. Cerebral Cortex 29(5):2211–27. doi: 10.1093/cercor/bhz026.
5. Salomons TV, Iannetti GD, Liang M, Wood JN (2016). The "Pain Matrix" in Pain-Free Individuals. JAMA Neurol 73(6):755-6. doi: 10.1001/jamaneurol.2016.0653.
6. Eklund A, Nichols TE, Knutsson H (2016). Cluster failure: Why fMRI inferences for spatial extent have inflated false-positive rates. PNAS 113(28):7900-7905. doi: 10.1073/pnas.1602413113.
7. Lewis LD, Setsompop K, Rosen BR, Polimeni JR (2018). Stimulus-dependent hemodynamic response timing across the human subcortical-cortical visual pathway identified through high spatiotemporal resolution 7T fMRI. Neuroimage 181:279-291. doi: 10.1016/j.neuroimage.2018.06.056.
Figures