3284
Automatic Prediction of MGMT and IDH Genotype for Gliomas from MR Images via Multi-task Deep Learning Network1Beijing Advanced Innovation Center for Big Data-Based Precision Medicine(BDBPM) ,Beihang University,100083, Beijing, China, 2Key Laboratory of Molecular Imaging, Institute of Automation, Chinese Academy of Sciences,100190, Beijing, China, 3Department of Radiology, First Clinical Medical College, Shanxi Medical University,030001, Taiyuan, China
Synopsis
In order to preoperatively predict the multiple genotype mutation for gliomas, we proposed an end-to-end multi-task deep learning model based on MR images analysis for simultaneously predicting IDH and MGMT mutation. Best-performed model was obtained by changing the number of sharing layers in the network, achieving accuracy of 79.78% for MGMT, 78.88% for IDH in the test dataset. Our results indicated that multi-task deep learning model provided a potential solution for simultaneously prediction of multiple genotype in gliomas.
Introduction
Isocitrate dehydrogenase (IDH) and O6-methylguanine methyltransferase (MGMT) genotype are both important genetic hallmarks with considerable prognostic value for gliomas1,2. Previous studies verified that certain radiological characteristics of MRI images were associated with the mutation status of MGMT and IDH 3,4. In this study, we constructed a multi-task learning model to simultaneously predict the mutation status of IDH and MGMT by analyzing magnetic resonance images (MRI).Materials and methods
MaterialsThis retrospective study was approved by the institutional review board of First Clinical Medical College, Shanxi Medical University. 187 gliomas patients receiving surgery operation and having status of IDH and MGMT genotype confirmed were enrolled in the current study. Preoperative MRI was performed with a 3.0-T scanner (Signa HDxt, GE Healthcare, USA) using an 8-channel array coil. The acquisition parameters were repetition time=220 ms, echo time=3.19 ms and slice thickness=4.5 mm. The region of interest (ROI) in the tumor area was manually segmented by 8 experienced neuroradiologists using ITK-SNAP (http://www.itksnap.org) .
Image preprocessing
A total of 2224 preoperative axial contrast enhanced T1-weighted images were assigned into training dataset and test dataset by the ratio of 8:2. Small patches containing ROIs were cropped from MRI images and resized into 96×96 pixels. Then, normalization5 were conducted on all the patches, followed by data augmentation to increase the dataset size to avoid overfitting.
Multi-task learning Model
Due to the small size of dataset, SmallerVGGNet structure with six convolution blocks was chosen as the initial model structure6. We developed a multi-task learning model with two branches that can predict the mutation status of IDH and MGMT respectively. Seven experiments for different number of sharing layers in the network were conducted to obtain the best-performed model. For each experiment, the sharing layers was defined respectively as n (n=0, 1, 2, 3, 4, 5, 6) blocks and the remaining layers was defined respectively as (6-n) blocks.
All convolution layers kernel was 3×3 and L2 regularization with weight 1×10-6 was used to avoid overfitting. The activation function of the output layer was sigmoid and Rectified Linear Units (ReLU) for other layers. The max pooling convolution core was 2×2. The dropout ratio in block6 was 0.5 and 0.25 for the others. The loss function consisted of 2 terms as shown in
$$$ Loss=α1∗Loss1 + α2∗Loss2$$$ (1)
Where α1 was 0.5 and α2 was 0.5. The loss of both branches used binary cross entropy, as shown in
$$$Loss1=−(y1∗log(ŷ 1)+(1−y1)∗log(1−ŷ 1))$$$ (2)
$$$Loss2=−(y2∗log(ŷ 2)+(1−y2)∗log(1−ŷ 2))$$$ (3)
Where y1 was MGMT mutation status, ŷ 1 was the probability of mutated MGMT, y2 was IDH mutation status, ŷ 2 was the probability of mutated IDH.
In the training step, the weights of layers were initialized by random numbers. The epoch was set as 200, and batch size was set as 32. Adam optimizer was initialized with a learning rate of 0.001 and decay of 5×10-5.
Results
The results are listed in Table 1. Comparing the results of the seven experiments, it can be observed that the model with sharing layers performed better than the model without sharing layers. Among the models with sharing layers, when the sharing layers was defined as 5 blocks and the remaining layers were 1 block the model performed best. The best-performed model structure was shown in Figure 1. Best-performed model achieved accuracy of 79.78% and AUC of 0.848 for MGMT, and accuracy of 78.88% and AUC of 0.863 for IDH in the test dataset on slice level.Conclusions and Discussions
The model with sharing layers performed better than that without sharing layers, indicating that multi-task learning provides a promising method when dealing with tasks having strong correlation with each other. The results of comparative experiments showed that the first several layers can be set as sharing layers to extract general features for prediction. Adjusting the number of sharing layers also provides a way to improve the performance of multi-task learning model.There are two limitations in this study. First, the sample size of the study is small, and this finding will be confirmed in a study with larger samples in the future. Second, the results are on slice level, our model will be used on patient level in the following study.
Acknowledgements
No acknowledgement found.References
[1] Brat DJ, V erhaak RG, Aldape KD et al. Comprehensive integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481-2498.
[2] Everhard S, Kaloshi G, Crinière E et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol.2006; 60(6):740-743.
[3]Drabycz S, Roldán G, de Robles P et al. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage. 2010; 49(2):1398-1405.
[4] Qi S, Y u L, Li H et al. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett. 2014; 7(6):1895-1902.
[5] Reinhold JC, Dewey BE, Carass A et al. Prince. Evaluating the impact of intensity normalization on MR image synthesis. Proc SPIE Int Soc Opt Eng.2019; 10949.
[6]Adrian Roserbrock, PyimageSearch, 07 May 2018. PyImageSearch-SmallerVGGNet: Multi-label classification with Keras. https://www.pyimagesearch.com/2018/05/07/multi-label-classification-with-keras/
Figures
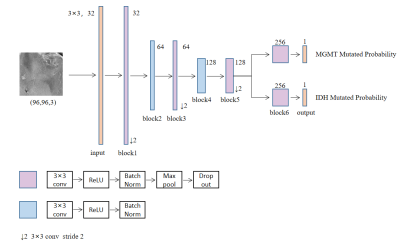
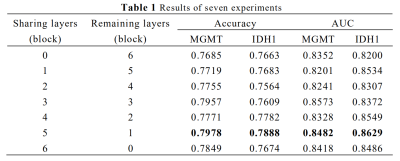
AUC, Area Under Receiver Operating Characteristic Curve
Sharing blocks means the number of blocks different branches owned jointly
Remaining blocks means the number of blocks different branches owned respectively