3279
Using MRI and Radiomics to Predict Pain in a Cohort of Trigeminal Neuralgia Patients Treated With Radiosurgery1Center for Magnetic Resonance Imaging, University of Minnesota, Minneapolis, MN, United States, 2Department of Neurosurgery, University of Minnesota, Minneapolis, MN, United States, 3Department of Diagnostic and Biological Science, University of Minnesota, Minneapolis, MN, United States
Synopsis
Due to a lack of objective markers, trigeminal neuralgia is difficult to diagnose and classify. This difficulty results in frequent pain recurrences following medication and invasive therapies. In this work, we show that a radiomics based model with features extracted from MRI images can be used to identify painful nerves. Using a cohort of trigeminal neuralgia patients, our predictive model achieves an accuracy of 78% and an AUC of 0.84 in distinguishing between nerves affected and nonaffected by trigeminal neuralgia.
Introduction
Trigeminal Neuralgia (TN) is a devastating chronic neuropathic condition that frequently requires invasive therapy. The diagnostic criteria for TN is primarily symptom based, and there are few, if any, objective markers1. This contributes to a lack of rational therapeutic guidance and a poor understanding surrounding the high rates of pain recurrence following intervention. Pain management in this style is inefficient and results in patients experiencing deleterious side effects and undergoing unnecessary invasive procedures2. MRI is typically used in the workup of trigeminal neuralgia patients to assess the presence and degree of neurovascular compression of the trigeminal nerve by nearby vascular structures. This remains a somewhat weak predictor of TN, because as much as a third of the healthy population also exhibits neurovascular compression based on MRI, whereas a large number of TN patients do not3. Thus, an objective biomarker for the classification of TN would help clinicians make treatment decisions. This work tests the hypothesis that radiomic features derived from MRI of the trigeminal nerve could be used in a predictive model to distinguish between TN afflicted nerves and their pain-free contralateral counterparts. The long term goal of this work is to develop a framework for automatic trigeminal neuralgia diagnosis and classification.Methods
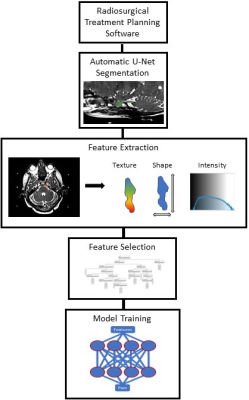
Treatment planning T1 and T2 weighted 1.5-Tesla MRI volumes were retrospectively acquired for patients undergoing Co-60 stereotactic radiosurgery to treat TN. Trigeminal nerves from the pons to the ganglion were segmented automatically by an in-house deep learning algorithm. Briefly, this algorithm consisted of a 3D UNet trained on a subset of the data. Segmentations were reviewed by hand to check if they (a) covered all of the trigeminal nerve from the pons to the anterior wall of Meckel’s cave and (b) did not overlap with any nearby brain tissue such as the pons or the temporal lobe. A total of 108 features consisting of image texture, shape, and intensity, were extracted from each imaging contrast for both nerves using the open source PyRadiomics package4. In order to mitigate overfitting, the top performing features were selected using a random-forest screening procedure, and a two layer shallow neural network was trained using the selected variables to differentiate between TN affected and non-affected nerves. The model was validated with five-fold cross validation.Results
A total of 134 patients (i.e. 268 nerves) were included in the predictive model. The average duration of pain prior to the procedure was 7 years. The top 20 performing features extracted from the masks were selected for the predictive model. The average validation set accuracy across the five-folds was 78%. The AUC of the model across the validation sets was 0.84, sensitivity and specificity were 0.82 and 0.76, respectively.Discussion
In general, diagnosing idiopathic TN from MRI is an intractable task to a human observer. Radiomics features perform well in distinguishing between pain-free and painful nerves in this dataset, suggesting that anatomical changes exist in painful nerves. However, a majority of the features included in the predictive model were based on image texture, so this information may not be easily ascertained by eye. This study is limited by the homogeneity of the patient population included. Namely, these are all patients whose pain is uncontrolled by medication and who lack a clear secondary cause for their trigeminal neuralgia. This work opens up the possibility of automatic detection and classification of TN. To achieve that, data from a wide variety of patient and pain types will need to be collected.Conclusion
Overall, this work suggests that image intensity and texture features from MR imaging of the trigeminal nerves are predictive of the presence of pain from TN. Further work will expand the training data to additional facial pain groups to boost the robustness of the model and explore the anatomical underpinnings of these findings.Acknowledgements
This research was supported by NIBIB P41 EB027061 and P30 NS076408. Personnel performing this research were also supported by the National Institutes of Health’s National Center for Advancing Translational Sciences, grants TL1R002493 and UL1TR002494. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences.References
1. Doshi TL, Nixdorf DR, Campbell CM, Raja SN. Biomarkers in Temporomandibular Disorder and Trigeminal Neuralgia: A Conceptual Framework for Understanding Chronic Pain. Can J Pain. 2020;4(1):1-18.
2. Zakrzewska JM, Akram H. Neurosurgical interventions for the treatment of classical trigeminal neuralgia. Cochrane Neuromuscular Group, ed. Cochrane Database Syst Rev. Published online September 7, 2011. doi:10.1002/14651858.CD007312.pub2
3. Magown P, Ko AL, Burchiel KJ. The Spectrum of Trigeminal Neuralgia Without Neurovascular Compression. Neurosurgery. 2019;85(3).
4. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77(21):e104-e107.