3275
Multi-layer backpropagation of classification information with Grad-CAM to enhance the interpretation of deep learning models1University of Calgary, Calgary, AB, Canada
Synopsis
Deep learning is becoming increasingly important in medical imaging analysis, but the ability to interpret deep learning models still lag behind. Here, based on a promising method, gradient-weighted class activation mapping (Grad-CAM), we developed new approaches to interpret arbitrary layers of a convolutional neural network (CNN). Further, using two common CNN models trained to classify brain MRI scans into 3 types, we demonstrated the promise of our new strategy. Characterizing features at low and high levels of a CNN may provide new biomarkers and new insight into disease mechanisms, deserving further validation.
Introduction
Deep learning algorithms have shown the potential to advance our disease characterization ability, but it remains a perplexity to how these deep learning models work. Several studies have highlighted the potential of heatmap-based techniques such as gradient-weighted class activation mapping (Grad-CAM)1 to interpretation the mechanisms of deep learning. However, most current strategies focus only on the last convolutional layer of a model. The interpretability and utility of the shallower layers are largely unknown. Our goal was to develop new techniques to explore the prospect of information backpropagation to arbitrary layers in a convolutional neural network (CNN) with Grad-CAM. Understanding the contribution of individual layers may have important implications for various biomarker studies in different diseases.Methods
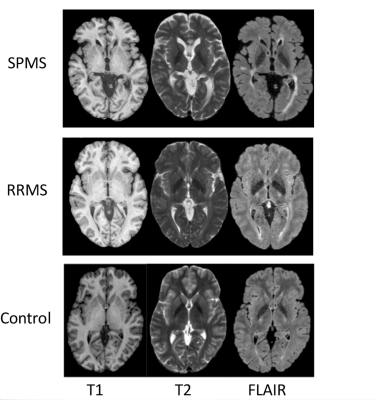
This study included two common CNNs pre-trained on ImageNet: VGG16 and VGG192. Using transfer learning, the architecture of the CNNs was modified to incorporate a global average pooling (GAP) layer prior to the output layer and then fine-tuned using brain MRI scans from 19 subjects with multiple sclerosis (MS) and 19 controls. Using T1, T2, and FLAIR MRI, the modified CNNs were optimized to classify the images into 3 types (Fig. 1): remitting MS (RRMS), secondary progressive MS (SPMS), or healthy control. The MR images were preprocessed through intra-subject co-registration (to T1), non-uniformity correction, and intensity normalization (0 to 1). In addition, the MR images that did not contain useful brain tissues at the very top and bottom of a head scan were removed, which resulted in 135 slices per sequence at training. The data were then augmented through rotation, translation, and scaling, and split into 3 portions: training (75%), validation (10%), testing (15%).The Grad-CAM algorithm used in this study started by finding the gradient of the final class score (yc) with respect to the feature maps, Ak of a specified layer. Weights were then calculated by taking the average of the newly computed gradients using the equation: $$$αck=1/zΣiΣj(αyc/αAkij)$$$, where i and j denote the width and height of the feature maps. Then the final Grad-CAM heatmap was calculated through a weighted sum with the following equation: $$LcGrad-CAM=ReLu(Σk αckAk)$$. ReLu was applied to eliminate any negative values thereby emphasizing positive contributions to the classification only. Finally, the heatmap was upsampled to the original size of the MRI and normalized.
The modified Grad-CAM algorithm was applied to all convolutional layers of the last block in the CNNs for heatmap comparisons. Furthermore, heatmaps were generated for a couple of additional layers prior to and following the last convolutional block of VGG19, the newer CNN method, to gain an additional perception of the interpretability of our new approach. Using Tesla K40m and V100 GPU, all analyses were conducted with the Keras deep learning framework and Tensorflow backend.
Results
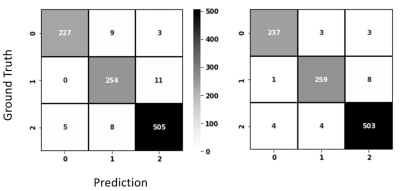
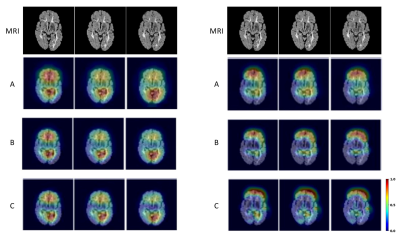
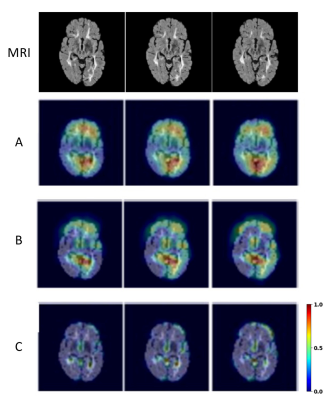
Both CNNs achieved relatively high accuracy in classifying the types of brain MRI scans as shown in the confusion matrices (Fig. 2). Grad-CAM backpropagation trials were conducted using brain MR images from a patient with SPMS, the type of disease known to have more severe tissue damage than other types (e.g RRMS). The heatmaps generated here from modifications to the backpropagating algorithm in Grad-CAM consistently highlighted distinct regions of activation (Fig. 2 & 3). While generalizability of the algorithm differed slightly between each iterative run of the different layers, results showed that backpropagation into the lower levels of the CNN layers resulted in greater localization, and backpropagation to higher levels showed greater generalization (Fig. 3). Further, additional tests with the VGG19 model illustrated that heatmaps resulted from backpropagation to the second last convolutional block in the CNN became difficult for humans to interpret as minimal regions of activation were highlighted (Fig. 4).Discussion
This study tested modifications of the Grad-CAM on two robust CNN classification models, VGG19 and VGG16 with GAP. The anatomical regions highlighting important features for classification were congruent with those shown in a previous study (manuscript under revision), where the frontal and temporal brain regions were specifically highlighted in SPMS pinpointing the importance of those regions in differentiating disease severity. Backpropagating to the last convolutional layer showed the best compromise between localization and generalization, consistent with the literature1, and the heatmap patterns appeared to be easier for human interpretation than other layers. The degree of generalization of heatmap patterns relates to the precision in identifying the most crucial regions of the brain underlying disease pathogenesis2, and different layers in a CNN contribute to different levels of abstraction of the extracted texture features in deep learning. Thus, assessing the contribution of features from individual layers may lead to new discoveries of biomarkers and disease mechanisms. In addition, further investigation of the critical brain areas indicated by different layers of Grad-CAM maps may stimulate identification of new therapeutic targets. Ultimately, these innovations should promote the clinical application of deep learning technologies.Conclusion
This novel feasibility study shows that it is possible to generate robust Grad-CAM maps from virtually any arbitrarily layer of a CNN through backpropagation of classification information. This approach would be critical to improve our model interpretation ability and disease understanding capacity in both MS and similar diseases.Acknowledgements
We thank the volunteers for participating in this study and the funding agencies for supporting this research: the Natural Sciences and Engineering Council of Canada (NSERC), Multiple Sclerosis Society of Canada, Alberta Innovates, Campus Alberta Neuroscience-MS Collaborations, and the HBI Brain and Mental Health MS Team, University of Calgary, Canada.References
1. Selvaraju, R. R., Cogswell, M., Das, A., Vedantam, R., Parikh, D., & Batra, D. (2017). Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. 2017 IEEE International Conference on Computer Vision (ICCV).
2. Ontaneda D and Fox RJ. Progressive multiple sclerosis. Curr Opin Neurol 2015; 28: 237-243. 2015/04/19.
Figures