3273
Early prediction of progression free survival and overall survival of patients with glioblastoma using machine learning and multiparametric MRI1Department of Radiology & Biomedical Imaging, University of California, San Francisco, SAN FRANCISCO, CA, United States, 2UCSF/UC Berkeley Graduate Program in Bioengineering, SAN FRANCISCO, CA, United States, 3Department of Neurological Surgery, University of California, San Francisco, SAN FRANCISCO, CA, United States
Synopsis
This study evaluates the predictive power of multi-parametric MRI at pre-therapy and mid-RT time points in predicting progression-free and overall survival of patients with glioblastoma (GBM). We trained and tested random forest models using metabolic, perfusion, and diffusion images at both preRT and midRT scans, and found that not confining these metrics to the anatomical lesion boundaries improved outcome prediction. The CEL volume mid-RT and type of treatment were among the most important features in predicting PFS, while the T2L volume and metabolic metrics at pre-RT were more relevant for OS prediction.
INTRODUCTION
Although glioblastoma (GBM) is a highly malignant and invasive brain tumor with poor overall survival (OS) of ~15 months, survival can vary substantially from less than 1 month to over 5 years [1]. OS is expected to be pushed further as newer antiangiogenic- and immuno-therapies, as well as precision-based approaches become available, bolstering the need for early prediction of individual prognosis before the onset of therapy to guide physicians in determining the appropriate personalized treatment strategy [2]. Recent studies have separately demonstrated the utility of metrics derived from metabolic, diffusion, and perfusion MRI at pre-, mid-, and post-radiotherapy time points as potential markers of clinical outcome of GBM patients [3-5]. In this study, we developed a machine learning model that incorporates multi-parametric metabolic and physiologic MRI from pre- and mid-therapy to predict OS and progression free survival (PFS) in patients with GBM using prospectively acquired data from 2 clinical trials evaluating angiogenic agents in combination with standard of care (SOC) radiation and temozolomide.METHODS
Subjects: This retrospective analysis included sixty-three patients with newly-diagnosed GBM and scanned after surgical resection, but before starting subsequent therapy. 28 patients received SOC treatment with radiation and temozolomide plus Avastin and Tarceva (ATT), while 35 patients received SOC treatment and Enzastaurin (ENZA). All patients underwent pre-RT baseline MRI, and 48 of these patients also received a mid-RT scan 3-4 weeks from the start of RT.Image Acquisition: MRI were performed on a 3T GE scanner using an eight-channel coil and consisted of: 1) standard anatomical imaging (T2-weighted FLAIR, 3D T1-weighted IR-SPGR pre- and post- the injection of contrast), 2) Diffusion-tensor imaging (DTI; b=1000 s/mm2, 6 directions, 4 excitations, TR/TE=1000/108ms, voxel size=1.7-2.0×1.7-2.0×2.0-3.0mm), 3) DSC perfusion-weighted imaging (T2*-weighted EPI, TR/TE/flip angle=1500/35ms/35°, 128×128 matrix, slice thickness=3mm, 80 time-points), 4) Lactate-edited 3D 1H-MRSI (flyback echo-planar readout in SI-direction, PRESS volume localization, VSS lipid suppression, excited volume=80×80×40mm, TR/TE=1100-1250/144ms, voxel-size=1×1×1cm).
Processing: All images and parametric maps were aligned to the T1-post-contrast images. Anatomical imaging was used to define the contrast-enhancing and T2-hyperintense lesions (CEL,T2L). Maps of apparent diffusion coefficient (ADC) and fractional anisotropy (FA) were derived from the diffusion imaging, while normalized peak height and relative cerebral blood volume maps (nPH and rCBV) were generated from the DSC-perfusion data using in-house software. From the metabolic data, maps of choline-to-NAA index (CNI), normalized lactate (nLac), and normalized lipid (nLip) were generated using NAA levels in normal-appearing white matter. Four different masks were created for the analysis: 1) CEL, 2) T2L, 3) supratentorial brain in PRESS box, and 4) T2L+(CNI>1).
Analysis: Descriptive statistics of parameters, including median, mean, percentiles (25 and 75), sum, kurtosis, and skewness, were derived from each parametric map. CEL and T2L volumes at both time points and type of treatment were also included in the feature space. Binary outcomes were split based on median PFS (45 weeks) and OS (76 weeks) of our cohort. A Mann–Whitney U-test was performed on the training data (44/63 patients) to determine relevant features to include in our machine learning model, with a threshold of p-value ≤ 0.1. For each mask, a random forest model was trained by performing 3-fold cross-validation on all imaging parameters from 44 patients, while 19 patients were withheld for testing. Each model was fit to maximize the area under the receiver operating characteristic curve (AUC-ROC).
RESULTS & DISCUSSION
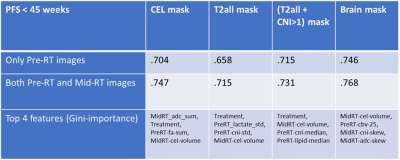
Figure 1 shows the pre-RT T2-FLAIR, nCBV, CNI, and nLac images of two patients with drastically different survival times. The visibly elevated CNI and rCBV near the T2L boundary in Patient A suggests a more aggressive lesion that resulted in shorter PFS and OS. Figure 2 shows box plots of the median values of each parametric map within the [T2L+(CNI>1)] mask. The only statistically significant parameters that differed between long and short survival groups were CEL volume at mid-RT and median CNI at pre-RT, suggesting that both pre-RT and mid-RT parameters may be useful in predicting outcome.Figures 3 and 4 display the performance of the Random Forest model on predicting long and short PFS and OS for each mask using just pre-RT images (first row), and both pre-RT and mid-RT images (second row). Predicting PFS was easier than OS, likely due to variations in subsequent therapy following initial progression. Using the brain mask cropped to the MRSI acquired volume on both pre-RT and mid-RT images gave the best ROC score of .768, supporting the notion that the most aggressive portion of these infiltrative tumors may lie outside of the anatomical lesion. The CEL mask gave the second-best performance when considering summed metrics. The CEL volume mid-RT and type of treatment (ATT vs. ENZA) were among the most important features in most PFS models, while the T2L volume and metabolic metrics at pre-RT were more relevant for OS prediction.
CONCLUSION
Our work highlights the importance of using multi-parametric MRI at pre-therapy and mid-RT time points in predicting PFS and OS, and not confining these metrics to the anatomical lesion boundaries. Although our results still have limited accuracy due to the small number of patients and specific therapies used, these findings can inform subsequent analyses of multiparametric MRI for predicting survival in patients with GBM.Acknowledgements
This research was supported by the NIH NCI grant P01CA118816References
(1) Marenco-Hillembrand, L.; Wijesekera, O.; Suarez-Meade, P.; Mampre, D.; Jackson, C.; Peterson, J.; Trifiletti, D.; Ham-mack, J.; Ortiz, K.; Lesser, E.; Spiegel, M.; Prevatt, C.; Hawayek, M.; Quinones-Hinojosa, A.; Chaichana, K. L. Journalof Neuro-Oncology 2020, 147, 297–307.
(2) Nelson, S. J.; Kadambi, A. K.; Park, I.; Li, Y.; Crane, J.; Olson, M.; Molinaro, A.; Roy, R.; Butowski, N.; Cha, S.; Chang,S. Neuro-Oncology 2017, 19, 430–439.
(3) Wen, Q.; Jalilian, L.; Lupo, J. M.; Li, Y.; Roy, R.; Molinaro, A. M.; Chang, S. M.; Prados, M.; Butowski, N.; Clarke, J.;Nelson, S. J. Translational Oncology 2015, 8, 446–455.
(4) Essock-Burns, E.; Lupo, J. M.; Cha, S.; Polley, M.-y.; Butowski, N. a.; Chang, S. M.; Nelson, S. J. Neuro-oncology 2011, 13, 119–131.
(5) Larsson, C.; Groote, I.; Vardal, J.; Kleppestø, M.; Odland, A.; Brandal, P.; Due-Tønnessen, P.; Holme, S. S.; Hope, T. R.;Meling, T. R.; Fosse, E.; Emblem, K. E.; Bjørnerud, A. Magnetic Resonance Imaging 2020, 68, 106–112.
Figures