3270
Automatic Vascular Function Estimation using Deep Learning for Dynamic Contrast-enhanced Magnetic Resonance Imaging1Radiology and Clinical Neuroscience, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 2Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, AB, Canada, 3Electrical and Computer Engineering, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 4General Electric Healthcare, Calgary, AB, Canada
Synopsis
Dynamic contrast-enhanced magnetic resonance (DCE-MR) imaging is an important clinical tool for investigating the cerebrovascular microcirculatory systems including the blood-brain barrier through estimation of perfusion and permeability maps. However, a vascular function is needed to generate these maps. The estimation is usually performed manually, potentially leading to error, and is also time-consuming. In this work, we designed a deep learning model that leverages the temporal and spatial information from a time series of DCE-MR images to estimate the vascular function automatically. Our model was able to generalize well for unseen data and achieved good overall performance.
Introduction
Dynamic contrast-enhanced magnetic resonance (DCE-MR) imaging is an important clinical and research tool for cancer imaging.1 DCE-MR imaging uses an injected contrast agent to investigate the microcirculation and the disruption of the blood-brain barrier by estimating perfusion and permeability maps. However, a necessary step for modeling the quantitative maps requires the extraction of a vascular function (VF).2 Commonly this step is performed manually in many DCE-MR analysis, making it both time-consuming and prone to human error. In the brain, both arteries and veins have been used to determine a VF.3,4,5 Here, we present a deep learning approach to automatically estimate a region over the transverse sinus on 4D DCE-MR images in order to estimate a VF. A unique property of this problem is that many regions in the vein yield similar vascular functions. Thus, the problem of estimating a VF can be simplified by finding a region over the vein that need not match the location of the manually selected region used to train the model.Materials and Methods
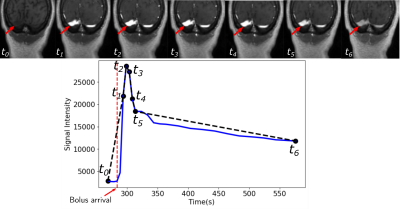
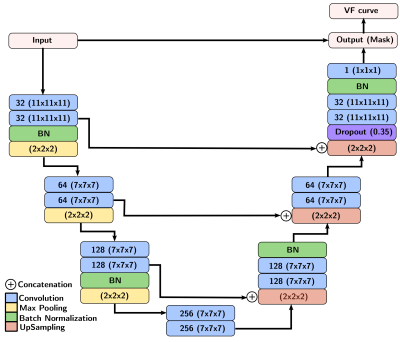
A total of 43 patients with glioblastoma were enrolled in this study (age: 59 ± 9.8 years [mean±standard deviation], 19 male). They were scanned post-resection using a protocol approved by our local research ethics board. A total of 155 baseline and follow-up DCE-MR image volumes were obtained from these patients. DCE-MR acquisition parameters were TR = 5 ms, TE = 1.9 ms, field-of-view of 240 mm × 240 mm, pixel size of 0.94 mm × 1.0 mm, and slice thickness of 2.0 mm. The DCE-MR image volumes were randomly divided into 100 (64.5%) volumes for training, 23 (14.8%) volumes for validation, and 32 (20.6%) volumes for testing. All images were normalized, on a per volume basis, by their maximum intensity to lie within the range [0,1]. The VF was extracted manually on the dynamic images by drawing a region of interest (ROI) on a coronal view containing the transverse sinus (Fig. 1). Because the VF had a priori defined characteristics (i.e., a sharp signal increase, followed by a short-duration maximum, and a slow decrease (Fig. 1)), the temporal dynamics of the data volume were undersampled for memory optimization. The VF sampling algorithm was based on the bolus arrival (BAT): One sample before the bolus arrival, five during bolus passage, and a seventh during the contrast wash-out (Fig. 2). The seven image volumes were then cropped to remove portions of the background that did not contribute to the VF and the temporal information was encoded into the seven channels of the input image.We used a 3D U-net architecture6 with three levels. The encoder consisted of repeated 11×11×11 and 7×7×7 padded convolutions, each followed by a leaky rectified linear unit (leaky ReLU,8 scale parameter=0.3), batch normalization, and a 2×2×2 max pooling operation. After each downsampling, the number of filters was doubled. The decoder consisted of upsampling and concatenation operation with successive 7×7×7 and 11×11×11 padded convolutions, each followed by a leaky ReLU, and a final layer 1×1×1 padded convolution followed by a sigmoid activation function (Fig. 3). As different regions over the transverse sinus can yield similar vascular functions, the predicted region does not need to match the location of the manually selected region. However, the separation between predicted and manual regions should be small. Thus, our loss function consists of two terms: a spatial term (distance between the predicted and manual region centers of mass, CoM, see coordinate definition in Fig. 4) and a temporal term (mean square error (MSE) between the sampled time signals). We evaluated the VF curve quality and how close our predicted region was to the manual region.
Results
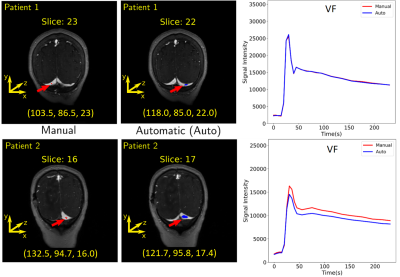
For training, validation, and testing we achieved MSE of 0.0028, 0.0035 and 0.0020, and CoM difference of 10.4, 5.4, and 11.4 voxel, respectively. The average absolute error in the CoM coordinates were (training: 9.12±8.16, 2.91±3.98, 1.57±1.65 voxels), (validation: 4.08±3.13, 1.68±1.66, 1.43±1.68 voxels), and (testing: 10.47±8.10, 2.4±2.61, 1.26±1.37 voxels). The overall prediction performance was good (Fig. 5). Examples of VF estimation for different patients are shown in Fig. 4.Discussion
The larger variance on the CoM x and y coordinates is due the fact that different regions over the transverse sinus can yield similar vascular functions, as shown in Fig. 5. Lower variance of the CoM z coordinate suggests that the predicted region was close to the slice of the manual region. The low MSE confirms that the predicted VF followed the same characteristics as the manual VF. Our VF prediction was close to the manual VF (Fig. 4).Conclusion
We demonstrated an automatic deep learning method can estimate a region over the transverse sinus in DCE-MR images can be used to identify a VF. Our method differs from other proposed deep learning methods,7 in that our model concurrently works with the spatial and temporal information, reducing the complexity of the network architecture. Our results demonstrated that our model generalizes well for unseen data and can be used to generate suitable VF. Next, we plan to compute and compare permeability maps using the manual and automatic vascular functions proposed in this work.Acknowledgements
University of Calgary - Eyes High Scholar Award.References
1. Winfield JM, Payne GS, Weller A, deSouza NM. DCE-MRI, DW-MRI, and MRS in Cancer: Challenges and Advantages of Implementing Qualitative and Quantitative Multi-parametric Imaging in the Clinic. Topics in Magnetic Resonance Imaging 25:245–254, 2016.
2. Calamante F. Arterial input function in perfusion MRI: A comprehensive review. Progress in Nuclear Magnetic Resonance Spectroscopy 74:1–32, 2013.
3. Bleeker EJ, Van Buchem MA, Van Osch MJ. Optimal location for arterial input function measurements near the middle cerebral artery in first-pass perfusion MRI. Journal of Cerebral Blood Flow and Metabolism 29:840–852, 2009.
4. Heye AK, Thrippleton MJ, Armitage PA, del C Valdes Hernandez M, Makin SD, Glatz A, Sakka E, Wardlaw JM. Tracer kinetic modelling for DCE-MRI quantification of subtle blood–brain barrier permeability. NeuroImage 125:446 – 455, 2016.
5. Ulas C, Das D, Thrippleton MJ, Valdes Hernandez MdC, Armitage PA, Makin SD, Wardlaw JM, Menze BH. Convolutional neural networks for direct inference of pharmacokinetic parameters: Application to stroke dynamic contrast-enhanced MRI. Frontiers in Neurology 9:1147, 2019.
6. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Lecture Notes in Computer Science, 9351:234–241, 2015.
7. Gordon Y, Partovi S, Müller-Eschner M, Amarteifio E, Bäuerle T, Weber MA, Kauczor HU, Rengier F. Dynamic contrast-enhanced magnetic resonance imaging: fundamentals and application to the evaluation of the peripheral perfusion. Cardiovascular diagnosis and therapy 4:147–14764, 2014.
8. Xu, Bing, Wang, Naiyan, Chen, Tianqi, Li, Mu. Empirical Evaluation of Rectified Activations in Convolutional Network, 2015.
Figures