3269
Automatic segmentation of arterial vessel wall on undersampled MR image using deep learning1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 2College of Software, Xinjiang University, Urumqi,, China, 3Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 4CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 5Shanghai United Imaging Healthcare Co., Ltd., shanghai, China, 6Department of Radiology, Guangdong Second Provincial General Hospital, guangdong, China
Synopsis
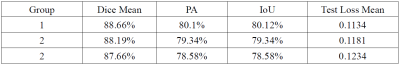
A total of 124 patients were included in this study. We used U-net neural network architecture to segment the arterial vessel wall on original acquired MR vessel wall images and the corresponding images reconstructed from undersampled K-space data. The Dice coefficients based on the original K-space data, the K-space data with a sampling rate of 7.7%, and K-space data with a sampling rate of 1.9% were 88.66%, 88.19%, and 87.66%, respectively. The effectiveness of arterial vessel wall segmentation on undersampled images using U-net network was verified. The result demonstrated the potential to improve the acceleration performance of MR imaging.
Introduction
Obtaining sufficient and accurate voxel information for patient diagnosis while scanning as little K-space data as possible is an eternal problem in MR imaging [1-2]. The significance of shortening the scanning time is to improve efficiency. Moreover, for patients who are difficult to maintain a static state, it is only possible to obtain MRI images by shorting the scanning time. Convolutional Neural Networks as a deep learning model has been widely used in medical image analysis. In the training, the data is divided into multiple groups, and the group is put into the network model for training one by one [3]. The parameters in the model can reflect the change process from input to output with training fine. Combining these methods, we can catch enough features from undersampled frequency through machine learning methods, achieve faster scanning, or simulates the function of segmentation on super-resolution images [4-5]. In this study, we developed and evaluated a U-net neural network architecture to segment the arterial vessel wall on original acquired MR vessel wall images and the corresponding images reconstructed from undersampled K-space data.Materials and Methods
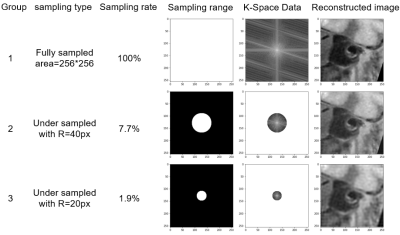
Data Acquisition: A total of 124 patients with intracranial atherosclerotic disease were included in the study. All patients underwent MR vessel wall imaging using a 3D MATRIX sequence on a 3-Tesla whole-body MR system (uMR 790, United Imaging Healthcare, Shanghai, China). The imaging parameters include: matrix size = 384x 320x256 , TR/TE = 800/13.92 ms, field of view =230x192x154 mm, ETL = 46, bandwidth (BW) = 600 Hz/pixel, uCS = 4.9. The annotation data used for training is generated manually by a radiologist.Data preprocessing:The experiment used two-dimensional fourier transform to transform medical images into the frequency domain [6-7]. These data in the frequency domain are called K-space data. First, the original acquired high-resolution images were converted into K-space data (called fully sampled K-space here) through Fourier transform. Then in the center of the fully sampled K-space data, a round box with different radius was used to undersamplethe fully sampled K-space. Finally, two different undersampled low-frequency areas were selected and converted into the image by inverse fourier transform to obtain two low-resolution image sets.
Training models: The study used the two image sets reconstructed from undersampled K-space data with different undersampled rates and the original images as different inputs to train the U-net network model for arterial vessel wall segmentation task. Figure 1 showed representative original images and undersampled images with different undersampled rate. Group 1 used fully sampled data of 100% sampling rate, group 2 used undersampled data of 7.7% sampling rate, and group 3 used undersampled data of 1.4% sampling rate. Deep learning segmentation was performed using the U-net, which is a fully connected convolutional residual network [8], and consists of convolution and max-pooling layers at the descending part (the left component of U). The segmentation performance was evaluated using the Dice rate, pixel accuracy (PA), and Intersection-over-Union (IoU).
Results
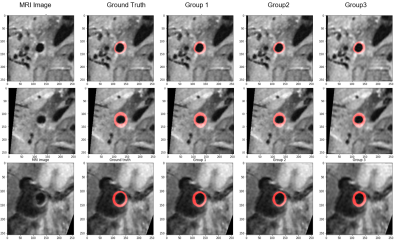
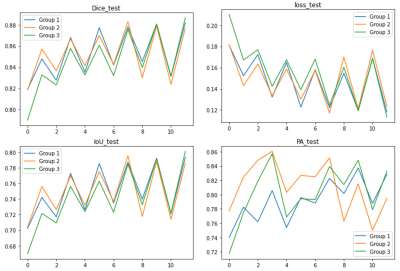
The Dice rate, PA, and IoU were calculated for each group respectively and summarized in Table 1. The Dice coefficients based on the original fully K-space data, the K-space data with a sampling rate of 7.7%, and K-space data with a sampling rate of 1.9% were 88.66%, 88.19%, and 87.66%, respectively. Figure 2 showed representative images of the segmentation results of different three group experiments. The correlation between the U-net prediction output and the training epoch for each group were shown in Figure 3-4. Among them, the Dice rate of sampling rate of 100% reached 80.77%, sampling rate of 7.7% reached 95.51%, and sampling rate of 1.9% reached 95.56%. The obtained results for three different groups were similar. To a certain extent reduce the sampling rate, the impact on the evaluation index is not significant.Discussion
The results showed that the use of undersampled data could also achieve similar results as using fully sampled data for segmentation using U-net training network. For the segmentation of arterial vessel wall based on undersampled images, reasonable accuracy segmented by using a template-based method compared to the ground truth was verified by a radiologist. The use of undersampled K-space data for the segmentation of arterial vessel walls has been verified as feasible and provided a new method for further acceleration of MRI. As the sampling rate of the image reduces, the performance did not change much, which might be in part due to the ground truth of different groups had the same accuracy. We could use higher resolution images for more accurate manual segmentation and simulated low-resolution images for deep learning training, then super-resolution segmentation results could be achieved. With the higher resolution, medical imaging equipment gradually becoming emergence can comprehensively improve the diagnostic accuracy of existing low-resolution equipment not limited by resolution, and the results may be used to develop a fully automatic diagnostic tool that can be implemented for clinical use.Acknowledgements
The study was partially support by National Natural Science Foundation of China (81830056), Natural Science Foundation of Guangdong Province (2018A030313204), and Shenzhen Basic Research Program (JCYJ20180302145700745 and KCXFZ202002011010360)。References
[1] Bao, Lijun et al. Undersampled MR image reconstruction using an enhanced recursive residual network. Paper presented at: Journal of magnetic resonance (San Diego, Calif.: 1997) vol.
[2] Hyun, Chang Min et al. Deep learning for undersampled MRI reconstruction. Paper presented at: Physics in medicine and biology vol. 63,13 135007. 25 Jun. 2018
[3] Mingxing Tan et al. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. Paper presented at: International Conference on Machine Learning, 2019.
[4] Bahrami, Khosro et al. 7T-guided super-resolution of 3T MRI. Paper presented at: Medical physics vol. 44,5 (2017): 1661-1677.
[5] Ebner, Michael et al. An automated framework for localization, segmentation and super-resolution reconstruction of fetal brain MRI. NeuroImage vol. 206 (2020): 116324.
[6] Ravishankar, Saiprasad et al. Image Reconstruction: From Sparsity to Data-adaptive Methods and Machine Learning. Paper presented at: Proceedings of the IEEE. Institute of Electrical and Electronics Engineers vol. 108,1 (2020): 86-109.
[7] Delfin, Leandro Morera et al. Driving Maximal Frequency Content and Natural Slopes Sharpening for Image Amplification with High Scale Factor. Paper presented at: Current medical imaging reviews vol. 16,1 (2020): 36-49.
[8] Ronneberger et al. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical image computing and computer-assisted intervention 2015.
Figures