3263
What we can learn from adults: Usability of two AI algorithms for Brain and tumor segmentation in a pediatric population.
Maxime DRAI1, GILLES BRUN1, Nadine GIRARD1,2, Benoit TESTUD1,3, and Jan-Patrick STELLMANN1,3
1Neuroradiology, APHM, Marseille, France, 2CRMBM-CEMEREM, Aix-Marseille Université, Marseille, France, 3CNRS, CRMBM-CEMEREM, UMR 7339, Aix-Marseille Université, Marseille, France
1Neuroradiology, APHM, Marseille, France, 2CRMBM-CEMEREM, Aix-Marseille Université, Marseille, France, 3CNRS, CRMBM-CEMEREM, UMR 7339, Aix-Marseille Université, Marseille, France
Synopsis
AI brain tumor segmentation and brain extraction algorithms are an essential step in image processing, however they are mainly developed in adults. Here, we aimed to explore the usability of these algorithms in a heterogenous pediatric population. In 42 brain pediatric tumor MRI, we compared manual mask with mask generated by the algorithms. Results were excellent for brain extraction, moderate for segmentation of contrast-enhancing tumors, and weak for non-enhancing T2-signal abnormalities. Some improvements are necessary to adapt this algorithm to pediatric brain tumors. However, borrowing strength from adults might be a feasible approach for AI implementation in rare pediatric populations.
INTRODUCTION
Many artificial intelligence (AI) tools in medical imaging have been created to support radiologists in daily practice1. Recently, automatic tumor segmentation2 and brain extraction[iii] algorithms, “HD-GLIOMA” and “HD-BET” (www.neuroAI-HD.org), have been developed and validated allowing a standardization in the follow-up of adult brain tumors and a better evaluation of the therapeutic response than the Response Assessment in Neuro-Oncology (RANO) criteria[iv]. However, this algorithm was trained from adults databases and only on gliomas. Standardized follow-up is also needed in pediatric populations, but case numbers are too low to easily train such algorithms.The purpose of this study is to assess these algorithms in a heterogenous population of pediatric brain tumors.METHODS
We screened our clinical database of pediatric brain tumors that had a complete pre-treatment examination with T1, T2, Flair and contrast-enhanced T1 (CT1). First, we used HD-BET to perform a brain extraction on all four sequences. In addition, we computed masks from FSL-BET (all sequences) and ROBEX (only T1) for comparison. We computed Dice coefficients between the different BET masks and a manual reference brain mask. In a second step, we used Dice coefficients to compare results from the tumor segmentation with HD-GLIOMA with manual masks for contrast-enhancing tumors (CE) and non-enhancing T2-signal abnormalities (NE) parts.RESULTS
We included 42 patients (25 males, 60%) with a median age of 6 years (range 0.6-17). Twenty-one (50%) of the tumors were classified as WHO grade4 IV tumors (medullosblastoma, glioblastoma, diffuse midline glioma) and six (14%) as WHO grade III tumors (anaspalatic ependydmoma, Pleomorphic xanthoastrocytoma), and 25 (59%) were located in the posterior fossa.Median Dice coefficient for brain extraction were overall lower for FSL-BET than for HDBET: 90.4 vs 99.6 for CT1, 95.0 vs. 99.8 for FLAIR, 75.1 vs. 98.9 for T1 and 89.4 vs 99.1 for T2 (Figure 1). The ROBEX algorithms performed also better than FSL, but slightly worse than the HD-BET. Median Dice coefficient for CE and NE were 78.2 and 24.4 respectively (Figure 2). Performance was better in the tumors located in the posterior fossa (83.0 and 45.4 for CE and NE respectively). Precision of CE masks was higher in low-grade tumors whereas NE masks of high-grade tumors tended to show a better overlap with expert masks.DISCUSSION
HD-BET provides a reliable extraction of the pediatric brain with different weighted images in 2D and 3D planes. HD GLIOMA works moderately for CE segmentation, especially in the infra-tentorial localization. However, NE segmentation has weak performance, partly due to the trans-ependymal resorption with secondary hydrocephalus that are more frequent than in adults. Moreover, our cohort included also a relevant number of non-glioma tumors, that the algorithm was never trained on. However, our results indicate that segmentation algorithms developed in adult populations might also be useful in pediatrics.CONCLUSION
Some improvements are necessary to adapt this algorithm to the special features of the pediatric brain tumors. Other studies with a larger dataset are needed to confirm our results.Acknowledgements
No acknowledgement found.References
- Filippo Pesapane, Marina Codari, et Francesco Sardanelli, « Artificial Intelligence in Medical Imaging: Threat or Opportunity? Radiologists Again at the Forefront of Innovation in Medicine », European Radiology Experimental 2, no 1 (24 octobre 2018): 35, https://doi.org/10.1186/s41747-018-0061-6.
- Philipp Kickingereder et al., « Automated Quantitative Tumour Response Assessment of MRI in Neuro-Oncology with Artificial Neural Networks: A Multicentre, Retrospective Study », The Lancet Oncology 20, no 5 (mai 2019): 728‑40, https://doi.org/10.1016/S1470-2045(19)30098-1.
- Fabian Isensee et al., « Automated Brain Extraction of Multisequence MRI Using Artificial Neural Networks », Human Brain Mapping 40, no 17 (2019): 4952‑64, https://doi.org/10.1002/hbm.24750.[1] Dewen Yang, « Standardized MRI assessment of high-grade glioma response: a review of the essential elements and pitfalls of the RANO criteria », Neuro-Oncology Practice 3, no 1 (1 mars 2016): 59‑67, https://doi.org/10.1093/nop/npv023.
- David N. Louis et al., « The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary », Acta Neuropathologica 131, no 6 (2016): 803‑20, https://doi.org/10.1007/s00401-016-1545-1.
Figures
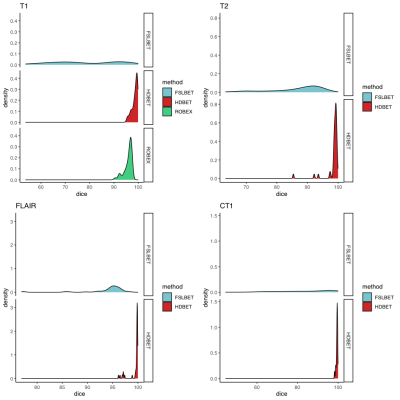
Figure 1 – Comparison of Dice coefficients between different brain extraction tools HD-BET and expert masks: Histograms showing the distribution of the values where values of 100% indicate e perfect overlap and 0 a complete lack of overlap.
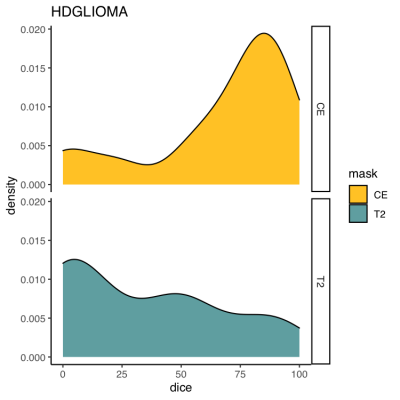
Figure 2 – Comparison of Dice coefficients between the tumor segmentation algorithm (HD-GLIOMA) and expert masks: Histograms showing the distribution of the values (100% indicates a perfect overlap and 0 a complete lack of overlap).