3262
Machine Learning Automatic Segmentation of Spinal Cord Lesions in Multiple Sclerosis Patients
Peter Hsu1, Sindhuja Govindarajan1, Nikhil Chettipally1, Lev Bangiyev2, Robert Peyster2, Giuseppe Cruciata2, Patricia Coyle2, Haifang Li2, Hasan Saffiudin1, Ryan Merritt1, Eric Wei1, Almighty Ironnah1, and Kwan Chen1
1Stony Brook University, Stony Brook, NY, United States, 2Stony Brook University Hospital, Stony Brook, NY, United States
1Stony Brook University, Stony Brook, NY, United States, 2Stony Brook University Hospital, Stony Brook, NY, United States
Synopsis
Multiple Sclerosis lesions in the spinal cord are associated with more debilitative disease outcomes and have predictive value for prognosis and diagnosis. However, these lesions are difficult to detect from MRI scans and this process is susceptible to inter-rater and intra-rater variability. Machine Learning techniques have the ability to assist in this problem. We propose a Convolutional Neural Network that can perform accurate identification and segmentation of MS lesions in the spinal cord. This method achieves high overlap with the segmentations of attending radiologists and is robust to imaging artifacts, showcasing the potential to be a tool for clinical practice.
Introduction
Multiple Sclerosis (MS) lesions in the spinal cord have been correlated with more aggressive MS and hold predictive value for disease diagnosis and prognosis1,4. The challenge for detecting and monitoring spine lesions is the high inter-rater and intra-rater variability from MRI scans. While several automated methods exist for MS lesions in the brain, there is only one existing software for the spine, Spinal Cord Toolbox (SCT)2. The purpose of this project was to develop an alternative method to automatically detect and segment MS lesions in the cervical spinal cord using a Convolutional Neural Network (CNN).Methods
After IRB approval, a retrospective PACS search was conducted to obtain 167 clinical MR images from MS patients. 1.5T and 3T Sagittal STIR images of the cervical spine from GE, Siemens, and Philips MRI machines were used for this study. These images followed a 2D MRI acquisition, 3400 repetition time, and 38.592 echo time. From this dataset, 147 were randomly separated into an 80/20 training/validation set and 20 were reserved for a testing set. Ground truth was created from the manual segmentations of five radiology residents which were validated by three attending radiologists. The ground truth for the testing set was formed by a consensus of three radiology residents and two attending radiologists.The spinal cord from each image was extracted using masks created by SCT. Image dimensions and voxel sizes varied considerably across images, requiring resampling to keep the data consistent. The height and length of images were resampled to 256x256 with the width remaining unchanged. Voxel sizes were resampled to an isotropic 1.0x1.0x1.0mm3. Linear contrast stretching was applied to reduce the internal variance of the images3. Data augmentation was also applied in the form of horizontal, vertical, and diagonal flipping.
The Machine Learning platform Tensorflow with Keras backend was used to adapt a 2D U-Net++ CNN architecture5. Our model consisted of 30 Convolutional Layers with batch normalization and max pooling to reduce overfitting. All layers utilized Exponential Linear Unit (ELU) activation functions except for the final layer which used a sigmoid activation function. The final layer predicted 0 or 1 for every pixel of a given image as being non-lesion or lesion, respectively.
Results
The primary metric for evaluation of image segmentation is the Dice Similarity Coefficient (DSC). Our model achieved a validation DSC of 0.6938 after training with a batch size of 2 across 100 epochs. On the 20 images reserved for testing, our model had a mean DSC of 0.6542. SCT was tested on the same data, achieving a mean DSC of 0.5375. The performance of three radiology residents was also recorded to compare the segmentations of our model to those of in-training radiologists. The other accuracy metrics consist of the Positive Predictive Value (PPV), Sensitivity, Specificity, False Positive Rate (FPR), False Discovery Rate (FDR), and False Negative Rate (FNR). The results across these accuracy metrics are summarized in Figure 1.Discussion
We have introduced a machine learning method for automatic detection and segmentation of MS lesions from the cervical spinal cord in MR images. By strict comparison on our dataset, our model outperforms the only alternative method, SCT, across all the segmentation accuracy metrics. These metrics were also used to compare the models to three radiology residents.Figure 2 showcases that our model is capable of identifying and segmenting MS lesions in the spinal cord. Our model achieves high overlap with the consensus ground truth, indicated by the high DSC score. The effectiveness of automated methods is highlighted in Figure 3. Both our model and SCT were able to successfully identify a lesion that none of the residents were able to. Our model is robust against motion artifacts in comparison to SCT and two of the three residents as highlighted in Figure 4. The performance benefits of our method give it the potential to serve as a useful tool for radiologists to quickly and accurately identify lesions in an image.
Despite the high performance of our model, further adjustments are needed. Radiology residents outperformed our model in mean PPV, mean specificity, mean FPR, and mean FDR. However, both specificity and FPR are highly influenced by the overwhelming number of true negatives in an MR image compared to false positives. Additionally, our dataset of 167 images is relatively small for training a CNN model, even with the use of data augmentation. Future studies would aim to expand this dataset to have hundreds or thousands of images. We also acknowledge the performance gains of our method compared to SCT can be attributed to utilizing our own dataset. A more apt comparison would require training across the same data and testing on an externally sourced set of data. Still, our method shows promising computational benefits utilizing a 2D architecture in comparison the 3D architecture used by SCT.
Conclusion
The use of CNNs can be efficient for automatic recognition and segmentation of spinal MS lesions in MR images. With a mean testing DSC of 0.6542, our model achieves competitive results compared to the only other software available for spinal cord lesion segmentation.Acknowledgements
No acknowledgement found.References
- Davda, N., Tallantyre, E., & Robertson, N. P. (2019). Early MRI predictors of prognosis in multiple sclerosis. Journal of Neurology, 266(12), 3171–3173. https://doi.org/10.1007/s00415-019-09589-2
- Gros, C., De Leener, B., Badji, A., Maranzano, J., Eden, D., Dupont, S. M., Talbott, J., Zhuoquiong, R., Liu, Y., Granberg, T., Ouellette, R., Tachibana, Y., Hori, M., Kamiya, K., Chougar, L., Stawiarz, L., Hillert, J., Bannier, E., Kerbrat, A., … Cohen-Adad, J. (2019). Automatic segmentation of the spinal cord and intramedullary multiple sclerosis lesions with convolutional neural networks. NeuroImage, 184, 901–915. https://doi.org/10.1016/j.neuroimage.2018.09.081
- Rafael C. Gonzales and Paul Wintz. (1987). Digital image processing 2nd Edition. Addison-Wesley Longman Publishing Co., Inc., USA.
- Sombekke, M. H., Wattjes, M. P., Balk, L. J., Nielsen, J. M., Vrenken, H., Uitdehaag, B. M. J., Polman, C. H., & Barkhof, F. (2013). Spinal cord lesions in patients with clinically isolated syndrome: A powerful tool in diagnosis and prognosis. Neurology, 80(1), 69–75. https://doi.org/10.1212/WNL.0b013e31827b1a67
- Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). UNet++: A Nested U-Net Architecture for Medical Image Segmentation. ArXiv:1807.10165 [Cs, Eess, Stat]. http://arxiv.org/abs/1807.10165
Figures
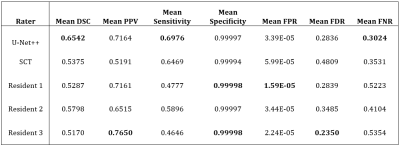
Comparison of the U-Net++, SCT, and three radiology residents on the 20
testing images of the cervical spinal cord. 15 images had lesions present and 5
had no lesions. Some control cases had other imaging artifacts to represent difficult or
uncertain cases. The best performance is highlighted in bold.
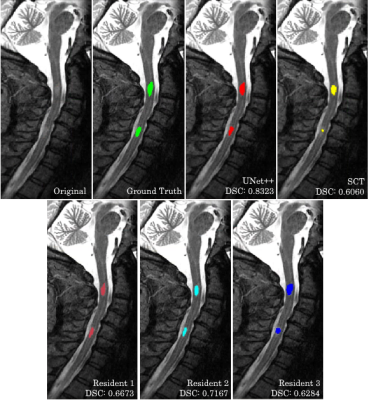
Segmentations made by our model, SCT, and three radiology residents in comparison to the consensus ground truth on an MR image of the spine with lesions. The DSC for this case is highlighted for each rater.
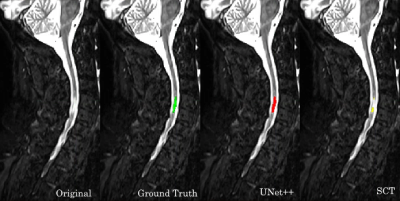
Segmentation performance of our model and SCT compared to the consensus ground truth. Their respective DSC scores were 0.7190 and 0.5814. The radiology residents did not manage to identify or segment this lesion.
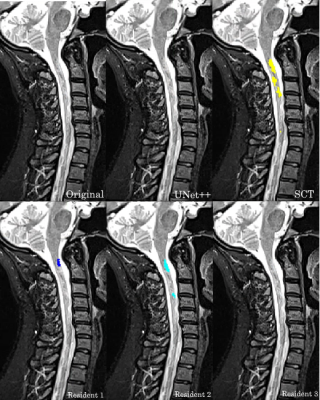
Performance of our model, SCT, and three radiology residents on a control case with motion artifacts present.