3261
A comparative study between multi-view 2D CNN and multi-view 3D anisotropic CNN for brain tumor segmentation1International Institute of Information Technology Bangalore (IIITB), Bengaluru, India
Synopsis
Brain tumor segmentation is one of the challenging image segmentation problem. Among the various architectures of CNN used for brain tumor segmentation from MRI images, we compare multi-view 2D CNN and multi-view 3D anisotropic CNN on the popular BraTS dataset. We computed four metrics to measure their performance on 10 test data. The first approach showed a mean sensitivity of 0.816 whereas second approach outperforms with mean sensitivity of 0.9217. Other metrics for both approaches achieved comparable results. Although both approaches consider all orthogonal planes, anisotropic CNN takes into account both global and local features efficiently thereby giving better results.
INTRODUCTION
MRI is an important tool to examine brain tumor. Over the last decade, various techniques from domains such as Computer vision and Machine learning have been studied for automated tumor segmentation in MRI images. While traditional methods are mostly semi-automated involving ROI detection, feature extraction and classification, several recent approaches are data-driven. Currently, Deep learning based fully-automated approaches are being extensively researched due to their unparalleled success across diverse tasks on natural images.The popular standard Neural networks that specialize in tasks such as segmentation, differ in several aspects, such as architecture, activation function, network depth, input dataset size, loss function and regularization. For the specific task of brain tumor segmentation, various techniques including CNN, RNN (Recurrent Neural Network), ensemble models have been studied. However, CNNs have shown the most prominent results 1. Diverse strategies to segment tumor from brain MRI employ 3D CNN in order to capture volumetric 3D features, however have the disadvantage of needing huge memory allocation. Approaches incorporating 2D CNN have low memory requirement but are unable to exploit the inherent 3D characteristics. Typically, the choice between using 2D CNN vs 3D CNN is based on the extent of balance required between receptive field, model complexity and memory consumption 2. Multi-path 2D CNN i.e. along each of the orthogonal axis (sagittal, coronal and axial) can exploit these features with low memory demand compared to 3D CNN 3. Commonly used methods to fuse predictions from the three axes are averaging, max pooling and majority voting 2,4-5. Besides, basic augmentation techniques for brain MRI images includes flip, rotation, scaling, cropping, translation, shearing, along with generating artificial data using GANs 6, 7.
The objective of the study is to examine the tumor delineation from the MR brain images. The focus is on being able to classify each of the brain voxels as belonging to either tumor-class or Non-tumor class. In this paper, we study the segmentation performance in two scenarios, namely (a) 2D CNNs which receive slice-wise inputs (b) 3D CNNS which receive brain volume inputs. The study is aimed at examining the following aspects: (a) Segmentation accuracy, as measured by standard metrics such as Dice score, as well as visual comparison of segmentation results (b) Computational complexity (resource requirement). Besides, it would also help answer if the brain volume which is inherently 3D data, can be processed as a stack of 2D images.
In this study we have utilized the popular BraTS dataset that is publicly available. The dataset comprises of four different MR contrasts, namely T1-weighted, T2-weighted, FLAIR, T1-contrast enhanced along with ground truth labels. Each of the modalities reveals some distinct tumor details and they all are significant to be considered. Using all four MR modalities, we have made a comparative study of two different approaches for the task of brain tumor segmentation.
METHOD
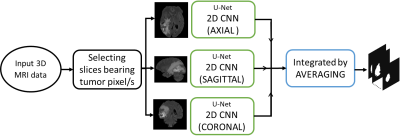
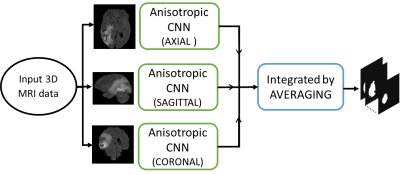
Our first approach have employed U-net architecture based three 2D CNNs, one for each sagittal, axial and coronal plane. Each model is trained on 259 data and tested on 10 data. Slices bearing tumor pixels are considered and rest are ignored thereby avoiding to train on unlabeled data and also save on training time. We have computed the tumor segmentation maps of 10 test data for each plane using 2D CNN and integrated them by averaging. The flow for this approach is illustrated in Fig. 1.In our second approach we have employed an architecture based on 3D anisotropic CNNs, one for each sagittal, axial and coronal plane 2. Because of the various drawbacks presented by 3D NNs, necessary tradeoffs between receptive field, complexity, memory consumption, etc. need to be considered. To employ these tradeoffs, the architecture uses Anisotropic and dilated convolutions, with larger receptive field in 2D and a relatively smaller receptive field in the orthogonal direction. The 3D kernel of size 3x3x3 is divided into two kernels, an intra-slice kernel (3x3x1) and an inter-slice kernel (1x1x3). The flow for this approach is illustrated in the Fig. 2. Adam optimizer is used for both the approaches with dice loss function.
RESULTS
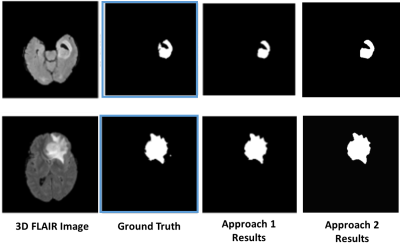
We implemented both our approaches on the Brats dataset and tested them on the data of 10 patients. The evaluation metrics are tabulated in Fig. 3. The segmentation results of two of the tested data are shown in Fig. 4.DISCUSSION AND CONCLUSION
In this study, we compared two approaches for the task of brain tumor segmentation; first approach utilized U-Net based on 2D CNN model while the other employed 3D Anisotropic CNN. Both the methods involved multi-view segmentation i.e. axial, sagittal and coronal planes and their results are fused by averaging. The segmentation results using first approach showed sensitivity of 81.6% while second approach give better sensitivity of 92.17%. However, the specificity, accuracy and dice score for both the approaches remains steady around 99.8%, 99% and 0.89 respectively. With these analyses, we can say that the method using 3D CNN gives better results than the method using 2D CNN, as it is able to extract global features along with the local features however at the cost of limited model complexity and high memory consumption.Acknowledgements
Ritu Lahoti and Lakshay Agarwal have contributed equally towards this work.References
1. Liu Z, Chen L, Tong L, Zhou F, Jiang Z, Zhang Q, et al. Deep Learning Based Brain Tumor Segmentation: A Survey. ArXiv. 2020;abs/2007.09479.
2. Wang GaL, Wenqi and Ourselin, Sébastien and Vercauteren, Tom. Automatic Brain Tumor Segmentation Using Cascaded Anisotropic Convolutional Neural Networks. Lecture Notes in Computer Science. 2018:178–90.
3. Wang G, Li W, Ourselin S, Vercauteren T. Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks With Uncertainty Estimation. Frontiers in Computational Neuroscience. 2019;13(56).
4. Banerjee S, Mitra S. Novel Volumetric Sub-region Segmentation in Brain Tumors. Frontiers in Computational Neuroscience. 2020;14(3).
5. Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Medical Image Analysis. 2018;43:98-111.
6. Nalepa J, Marcinkiewicz M, Kawulok M. Data Augmentation for Brain-Tumor Segmentation: A Review. Frontiers in computational neuroscience. 2019;13:83-.
7. Myronenko A. 3D MRI brain tumor segmentation using autoencoder regularization2018.
Figures