3256
Outcome prediction in Mild Traumatic Brain Injury patients using conventional and diffusion MRI via Support Vector Machine: A CENTER-TBI study1Radiology, UZA - Antwerp University Hospital, Antwerpen, Belgium, 2imec-Vision Lab, University of Antwerp, Antwerpen, Belgium, 3BioMedIA Group, Department of Computing, Imperial College London, London, United Kingdom, 4Division of Anaesthesia, Department of Medicine, University of Cambridge, Cambridge, United Kingdom, 5MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge, United Kingdom, 6Clinical Sciences, Diagnostic Radiology, Lund University, Lund, Sweden, 7Physiopharmacology, University of Antwerp, Antwerpen, Belgium
Synopsis
Over 40% of patients after a mild traumatic brain injury (mTBI) may have persisting symptoms. This study investigated a Support Vector Machine (SVM) approach for outcome prediction after mTBI from multi-modal MRI. The datasets included 77 mTBI patients from the CENTER-TBI study with acute T2w, SWI, FA and MD scans and outcome scores six month post-injury. Benefits of data harmonization were tested and Z-scoring reduced site-specific biases yielding 67.7% prediction accuracy. Our data-driven approach revealed that predictive signal was retrieved mainly from diffusion maps rather than conventional images, and was located in the superior fronto-occipital fascicle and the corticospinal tract.
Introduction
Mild traumatic brain injury (mTBI) accounts for over 85% of head injuries, and affects millions worldwide each year. In ~40% of the cases, recovery may be incomplete, with patients having persistent motor/psychological impairment for months to years after injury.1,2 Previous studies have been able to differentiate patients from controls. However, it remains a significant challenge in the acute setting to predict which patients will have good or poor recovery.3,4 In this study, we present results of outcome prediction after mTBI by means of a Support Vector Machine (SVM) based on conventional and diffusion MRI from the large-scale, multi-site CENTER-TBI study.5Methods
We included acute MRI scans of 77 mTBI patients from CENTER-TBI (within 7 days of injury, age: 17-70 years, Glasgow Coma Scale: 13-15). Patients were dichotomized according to their extended Glasgow Outcome Scale (GOSE) scores at six months; good (42 patients, GOSE=8) and poor (35 patients, GOSE<8) outcome. Data was acquired with semi-harmonized MR protocols6 at seven sites across Europe. T2-weighted (T2w), susceptibility weighted images (SWI), fractional anisotropy (FA) and mean diffusivity (MD) maps were coregistered to T1w images, which were non-linearly registered to a template, and then all images were directly projected to template space (applying transformation from coregistation and non-linear registration at once). The data was harmonized via Z-scoring, scanner-wise standard-scaling by the control data (126, age: 19-32 years). SVMs were either trained on the unharmonized or Z-scored harmonized datasets to predict patient outcome in a 4-fold cross-validation. Next, recursive feature elimination (RFE)7 was used to find the 10% most discriminative white matter voxels between both patient groups. Voxels identified in at least two folds were mapped to the JHU ICBM-DTI-81 atlas8 to determine their location within the brain. The voxels that yielded the highest accuracy were further examined by comparing their intensities across patient groups and control subjects via one-way ANOVA.Results
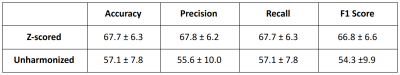
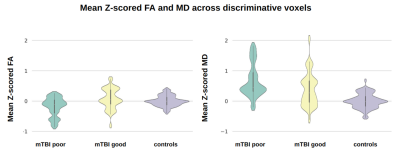
SVMs performed better when trained on Z-scored FA/MD/T2w/SWI, compared to unharmonized data (Table 1). The Z-score SVM predicted mTBI good and poor outcome with a mean accuracy of 67.7 ± 6.3% across the 4 folds.Z-scored voxels that remained after RFE were mostly selected from diffusion maps (FA: 30%, MD: 40%) rather than conventional images (T2w: 24%, SWI: 6%). Figure 1 shows the mean intensity across the select Z-scored FA and MD voxels for the mTBI good and poor outcome and healthy controls. For the Z-scored FA and MD selected voxels, the differences between groups is statistically significant (p<0.05 mTBI good vs mTBI poor and p<0.001 mTBI good/poor vs controls), yielding a good metric for outcome prediction among those groups.
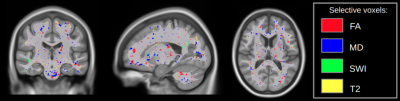
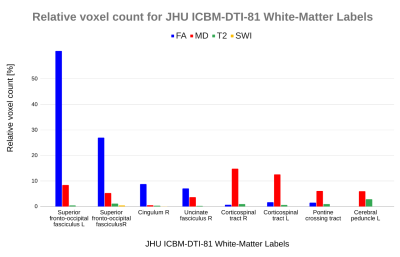
Locating the Z-scored selected voxels revealed various regions with a high number of predictive voxels, such as the superior fronto-occipital fasciculus for FA, corticospinal tract for MD, corona radiata for T2w and sagittal stratum for SWI (Figures 2 and 3).
Discussion
The prediction of outcome of mTBI patients is challenging and complex, especially given that the majority of patients have normal conventional neuroimaging. This study showed that FA and MD may provide information for outcome prediction in a multi-modal machine learning approach. Most discriminative voxels were selected from FA and MD maps, while T2w and SWI, which are usually used in clinical assessments, were less decisive for outcome prediction.Z-scoring was calculated site/scanner-specifically for each image modality, removing site-biases and yielding a higher accuracy. Moreover, the statistical analysis of Z-scored FA and MD (Figure 1) showed significant differences between poor and good outcome patients, suggesting that the selected voxels might represent a microstructural deterioration related to the trauma, which can be used to predict outcome. Previously, outcome prognosis has been investigated by many single-site MRI studies, however, no statistical significance was demonstrated across mTBI groups.4,9
The prediction was obtained for a specific set of selected voxels, which represent known structures that were mapped according to the JHU ICBM-DTI-81 atlas. FA voxels associated with outcome were predominantly sited on the superior fronto-occipital fasciculus, cingulum and uncinate fasciculus, while for MD it was mainly on the corticospinal tract, pontine crossing tract and cerebral peduncles. Predictive T2w voxels were identified on the posterior and anterior radiatas, superior longitudinal fasciculus and cerebral peduncles, in contrast to SWI that had few selected voxels mainly on the sagittal stratum (Figures 2 and 3). Additionally, the regions identified in the current study have been associated with mTBI outcome in previous single site studies.10
Conclusion
Predictive models capable of identifying mTBI patients with potential poor outcome could facilitate diagnosis and therapeutic decisions. This study showed the potential of SVM to predict patient outcome based on acute diffusion MRI. When combined with RFE and Z-scoring a prediction accuracy of 67.7% was obtained. The anatomical regions that were discriminative for mTBI outcome prediction were previously identified in single-site studies. We replicate these findings in a multi-center, multi-sequence study, and demonstrate the potential utility of voxel-wise mTBI outcome prediction using SVMs combined with recursive feature elimination and data harmonization.Acknowledgements
Research Grants: B-Q MINDED (EU H2020 764513) and CENTER-TBI (EC 602150)References
1-Galgano, M., Toshkezi, G., Qiu, X., Russell, T., Chin, L., & Zhao, L.-R. (2017). Traumatic Brain Injury. Cell Transplantation, 26(7), 1118–1130. https://doi.org/10.1177/0963689717714102
2-Carroll LJ, Cassidy JD, Peloso PM, Borg J, von Holst H, Holm L, et al (2004). Prognosis for mild traumatic brain injury: results of the WHO collaborating centre task force on mild traumatic brain injury. J Rehabil Med, 36(43), 84–105. https://doi.org/10.1080/16501960410023859
3-Lingsma, H. F., Yue, J. K., Maas, A. I. R., Steyerberg, E. W., Manley, G. T., Cooper, S. R., Dams-O’Connor, K., Gordon, W. A., Menon, D. K., Mukherjee, P., Okonkwo, D. O., Puccio, A. M., Schnyer, D. M., Valadka, A. B., Vassar, M. J., & Yuh, E. L. (2015). Outcome Prediction after Mild and Complicated Mild Traumatic Brain Injury: External Validation of Existing Models and Identification of New Predictors Using the TRACK-TBI Pilot Study. Journal of Neurotrauma, 32(2), 83–94. https://doi.org/10.1089/neu.2014.3384
4-Yuh, E. L., Cooper, S. R., Mukherjee, P., Yue, J. K., Lingsma, H. F., Gordon, W. A., Valadka, A. B., Okonkwo, D. O., Schnyer, D. M., Vassar, M. J., Maas, A. I. R., Manley, G. T., Casey, S. S., Cheong, M., Dams-O’Connor, K., Hricik, A. J., Inoue, T., Menon, D. K., … Morabito, D. J. (2014). Diffusion Tensor Imaging for Outcome Prediction in Mild Traumatic Brain Injury: A TRACK-TBI Study. Journal of Neurotrauma, 31(17), 1457–1477. https://doi.org/10.1089/neu.2013.3171
5-Maas, A. I. R., Menon, D. K., Steyerberg, E. W., Citerio, G., Lecky, F., Manley, G. T., Hill, S., Legrand, V., & Sorgner, A. (2015). Collaborative European NeuroTrauma Effectiveness Research in Traumatic Brain Injury (CENTER-TBI). Neurosurgery, 76(1), 67–80. https://doi.org/10.1227/neu.0000000000000575
6- MRI Study Protocols - CENTER-TBI. https://www.center-tbi.eu/project/mri-study-protocols
7- Guyon, I., Weston, J., Barnhill, S., & Vapnik, V. (2002). Gene selection for cancer classification using support vector machines. Machine Learning, 46(1/3), 389–422. https://doi.org/10.1023/a:1012487302797
8- Mori, S., Oishi, K., Jiang, H., Jiang, L., Li, X., Akhter, K., Hua, K., Faria, A. V., Mahmood, A., Woods, R., Toga, A. W., Pike, G. B., Neto, P. R., Evans, A., Zhang, J., Huang, H., Miller, M. I., van Zijl, P., & Mazziotta, J. (2008). Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. NeuroImage, 40(2), 570–582. https://doi.org/10.1016/j.neuroimage.2007.12.035
9- Hellstrøm, T., Kaufmann, T., Andelic, N., Soberg, H. L., Sigurdardottir, S., Helseth, E., Andreassen, O. A., & Westlye, L. T. (2017). Predicting Outcome 12 Months after Mild Traumatic Brain Injury in Patients Admitted to a Neurosurgery Service. Frontiers in Neurology, 8. https://doi.org/10.3389/fneur.2017.00125
10- Narayana, P. A. (2017). White matter changes in patients with mild traumatic brain injury: MRI perspective. Concussion, 2(2), CNC35. https://doi.org/10.2217/cnc-2016-0028
Figures