3249
Deep Learning Approach for Lumbosacral Plexus Segmentation from Magnetic Resonance Neurography: Initial Study1Department of Medical Imaging, The Third Affiliated Hospital of Southern Medical University, Guangzhou, China, 2Guangdong Provincial Key Laboratory of Medical Image Processing, Southern Medical University, Guangzhou, China, 3School of Biomedical Engineering, Southern Medical University, Guangzhou, China, 4China International Center, Philips Healthcare, Guangzhou, China
Synopsis
Accurate regional segmentation of the Lumbosacral Plexus (LSP) on magnetic resonance neurography (MRN) images is a fundamental requirement before LSP related disorders diagnosis can be achieved. In this paper, we utilize U-Net to segment LSP trunk and branch from three-dimensional fast field echo(3D-FFE) with principle of selective excitation technique (Proset) images. The results show that a U-Net deep learning framework expresses highly performance and less time-consumption for LSP segmentation in patients with degenerative spinal diseases and healthy subjects.
Purpose and Introduction
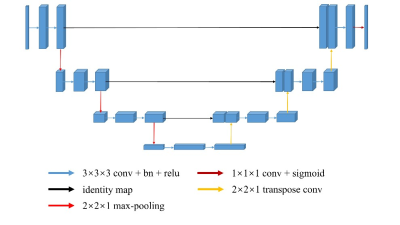
The lumbosacral plexus comprises a network of nerves, severe lumbosacral plexopathy could cause a serious lower extremity dysfunction and even permanent disabling condition.1 Magnetic resonance neurography(MRN) plays the most effective role in noninvasively identifying the type, location, and scope of lumbosacral plexus disorders and their underlying causes compared with clinical findings and electrodiagnostic test results2, 3. Accurate localization and identification of lumbosacral plexus are helpful for the diagnosis of diseases. Deep learning based on convolution neural networks has got popularity in medical field because of its powerful ability and has achieve excellent results in recognition and segmentation of normal anatomical structures and lesions. U-Net is a specialized CNN and has shown effectiveness in biomedical image segmentation due to its quality of easily integrate multi-scale information4, 5. The aim of this study was to determine the feasibility of using a deep learning approach to segment lumbosacral plexus within the coronal three-dimensional fast field echo (3D-FFE) with principle of selective excitation technique (Proset) sequence.Materials and Methods
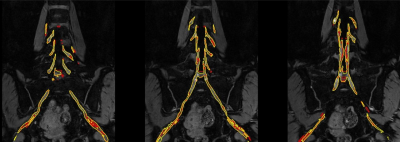
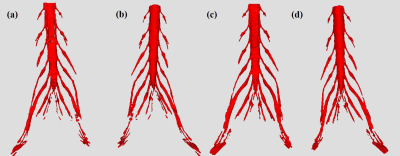
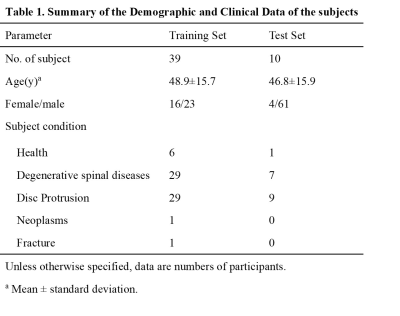
For this retrospective study, a total of forty-nine subjects of lumbosacral plexus magnetic resonance neurography images were obtained on a clinical whole-body 3.0T MRI system (Ingenia 3.0 T; Philips Health care, Best, the Netherlands). All MRN images were manually segmented for lumbosacral plexus using Mimics Medical software (version 21.0) by a junior radiologist with 5 years of experience. There were 39 and 10 volumes for training and test sets. A 3D U-Net was trained to extract lumbosacral plexus from the MRN images and five-fold cross validation was used to evaluation the performance of model. In inference phase, in order to get a better result, these models of five folds are all used for single image and the final segmentation mask is gained by averaging all outputs of five models. The Dice Similarity Coefficient (DSC), Sensitivity (SEN), and Positive Predictive Value (PPV) were used to evaluate the performance compared to manual segmentation. In addition, the segmentation time was manually segmented by a radiologist and automatically segmented by the U-Net model on a randomly chosen group of 10 cases.Results
Radiographs obtained to segment lumbosacral plexus in 49 subjects (mean age,48.49 years ±15.58[standard deviation]; 29 men) with or without lumbar intervertebral disc protrusion and bone neoplasms. Manual segmentation takes approximately 2 hours and 19 minutes per subject. Automatic lumbosacral plexus segmentation of U-Net takes less than 1 second, which was significantly faster than manual segmentation. The Dice, SEN, and PPV of the U-Net model for segmenting was 0.855±0.017, 0.848±0.041, and 0.864±0.017, respectively.Conclusion
A fully automatic U-net model was able to segment lumbosacral plexus accurately and shorten the segmentation time significantly.Acknowledgements
Jian Wang and Guohui Ruan contributed equally to this work.
Xiaodong Zhang and Yanqiu Feng are both co-corresponding authors.
Funding: The National Natural Science Foundation of China (81801653, 81871349, and 61671228), Science and Technology Planning Project of Guangdong Province (2017B090912006) .
References
1.Demystifying MR Neurography of the Lumbosacral Plexus: From Protocols to Pathologies. BioMed research international 2018; 2018: 9608947.
2.Eppenberger P, Andreisek G, Chhabra A. Magnetic resonance neurography: diffusion tensor imaging and future directions. Neuroimaging clinics of North America 2014; 24(1): 245-256. 3.Chhabra A, Lee PP, Bizzell C, Soldatos T.
3 Tesla MR neurography--technique, interpretation, and pitfalls. Skeletal radiology 2011; 40(10): 1249-1260.
4.Livne M, Rieger J, Aydin OU, Taha AA, Akay EM, Kossen T et al. A U-Net Deep Learning Framework for High Performance Vessel Segmentation in Patients With Cerebrovascular Disease. Frontiers in neuroscience 2019; 13: 97.
5.Zabihollahy F, Schieda N, Krishna Jeyaraj S, Ukwatta E. Automated segmentation of prostate zonal anatomy on T2-weighted (T2W) and apparent diffusion coefficient (ADC) map MR images using U-Nets. Medical physics 2019; 46(7): 3078-3090.
Figures