3224
Generalizable deep learning for multi-resolution proton MRI lung segmentation in multiple diseases1POLARIS, University of Sheffield, Sheffield, United Kingdom, 2Oncology and Metabolism, University of Sheffield, Sheffield, United Kingdom
Synopsis
We evaluate a fully-automated generalizable deep learning (DL) approach for lung segmentation using a 3D convolutional neural network on a large and diverse proton (1H) MRI dataset, containing images acquired at different resolutions and inflation levels. The dataset comprised of 336 1H-MR images from healthy subjects and patients with respiratory diseases. Our trained model was able to accurately segment scans of markedly different resolutions (3x3x3mm3, 4x4x5mm3 and 4x4x10mm3), achieving a mean±SD Dice similarity coefficient of 0.94±0.02. In addition, it was shown that DL generates more accurate segmentations compared to state-of-the-art solutions.
Introduction
Accurate segmentations of the lung parenchyma from thoracic 1H-MRI scans are crucial for several pulmonary applications, including the computation of thoracic cavity volume required for ventilated defect percentage1 and the generation of contrast-free surrogates of lung function2. However, previous methods such as spatial fuzzy c-means (SFCM)3 are semi-automatic and require significant time from experienced observers for manual editing. Moreover, such algorithms do not perform well across different acquisition protocols. Recently, deep learning (DL) has shown great promise for numerous segmentation problems.4 Here, we propose a generalizable deep learning approach to 1H-MRI lung segmentation using a 3D convolutional neural network (CNN). We assess the performance of the network across multiple resolutions, breathing manoeuvres and diseases. Furthermore, we compare DL performance with that of SFCM.Methods
Imaging dataWe retrospectively pooled 334 1H-MRI scans from 79 subjects who were healthy volunteers (n=14) or patients with cystic fibrosis (CF) (n=24), asthma (n=23) or lung cancer (n=18).
MRI acquisition
All subjects underwent 3D spoiled gradient-recalled echo (SPGR) 1H-MRI with full-lung coverage at 1.5T. One of three acquisition protocols were used which yielded the following resolutions: 1) 3x3x3mm3; 2) 4x4x5mm3; 3) 4x4x10mm3. Lung inflation levels included residual volume, functional residual capacity and total lung capacity. Figure 1 shows example coronal slices for the three different acquisitions for a single CF patient.
DL segmentation
A CNN with a 3D UNet5 architecture was trained with a PreLU activation function, ADAM optimisation and cross-entropy loss function using NiftyNet6. A learning rate of 0.00001 and batch size of 2 was used. A decay of 1x10-6 and L2 regularisation were selected to minimize overfitting. The model was trained using 304 scans (see Figure 2). 10% of the training data was used for internal validation. The testing set comprised of 30 scans, 10 from each of the three acquisition procedures. Although longitudinal and multi-inflation level scans were included, no patient was present in both the training and testing sets.
Data analysis
To evaluate segmentation accuracy, Dice similarity coefficients (DSCs), Average Boundary Hausdorff distance (Avg-HD), 95th percentile Hausdorff distance (HD95) and XOR7 metrics were computed. Mann-Whitney tests were used to assess group differences between the three acquisitions. Pearson correlation and Bland-Altman analyses were conducted to compare the volumes of the DL and expert segmentations. The DL segmentations were compared with those generated by an established SFCM method.3
Results
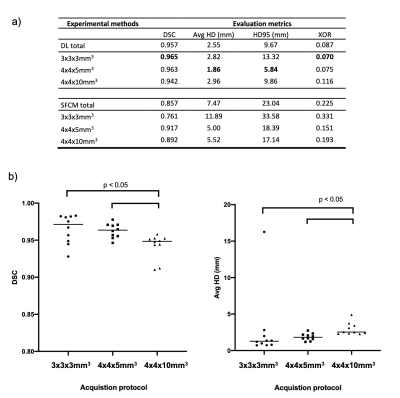
Figure 3 shows the qualitative and quantitative performance of DL and SFCM segmentations for three cases with different resolutions and diseases; DL segmentations accurately depict those rendered by an expert, avoiding major airways and following the lung boundaries accurately. A full breakdown of results for each acquisition protocol is displayed in Figure 4a. DL achieved a median DSC>0.94 and Avg-HD<2.5mm over all scans. The highest performance is shown for the 3x3x3mm3 scans using the DSC and XOR metrics, whilst the 4x4x5mm3 scans demonstrate the best performance using the Avg-HD and HD95 metrics. DL outperformed the conventional SFCM method across all metrics and acquisitions.Figure 4b shows the DL results for the three resolutions in the testing set; no statistically significant difference was observed between the 3x3x3mm3 and 4x4x5mm3 scans (p>0.05), although the segmentations generated on both resolutions outperformed those of the 4x4x10mm3 scans (p<0.05).
Correlation and Bland-Altman analyses of lung volumes for DL and SFCM segmentations against the expert segmentations are shown in Figure 5. DL exhibited strong correlation (r=0.99; p<0.0001) and limited bias of -0.015 litres, indicating that the algorithm contains no bias towards predicting smaller volumes and no trend in diminishing accuracy as the volume increases or decreases, is observed. In contrast, the SFCM method exhibited significantly increased bias of 0.84 litres and weaker correlation (r=0.90; p<0.0001). The DL method generated segmentations in approximately 30 seconds compared to 2 minutes for SFCM.
Discussion
By training on a dataset that contains a wide range of acquisition protocols, diseases and inflation levels, the CNN was able to generate accurate lung segmentations that generalized well across different scans. However, the segmentation accuracy reduced for the 4x4x10mm3 scans. This is potentially due to an imbalance in the proportion of these scans in the total dataset; out of 333 scans, only 68 were acquired at this resolution with the remainder divided almost equally between the other two resolutions. Consequently, the dataset was unevenly distributed in favour of higher resolution scans. In future work, we will address this imbalance to further increase the accuracy of DL segmentations for lower resolution scans.Whilst it was ensured that no patient was included in both the training and testing sets, scans at multiple inflation levels or time points from the same patient were included in either the training or testing sets. Therefore, there could be biases towards patients with the most scans in the dataset; in most cases, however, the number of scans per patient was similar, thereby reducing the impact of such biases.
Conclusion
In this study, we evaluated a 3D CNN that yields highly accurate lung segmentations and is robust to image resolution, breathing manoeuvre and disease. The CNN significantly outperformed the SFCM method and is expected to eliminate or dramatically reduce the time taken to generate and manually edit lung segmentations.Acknowledgements
This work was supported by Yorkshire Cancer Research, Weston Park Cancer Charity, National Institute of Health Research and the Medical Research Council.References
1. Woodhouse N, Wild J.M, Paley M, et al. Combined helium‐3/proton magnetic resonance imaging measurement of ventilated lung volumes in smokers compared to never‐smokers. J. Magn. Reson. Imaging, 2005;21:365-369.
2. Bauman G, Puderbach M, Deimling M, et al. Non-contrast-enhanced perfusion and ventilation assessment of the human lung by means of fourier decomposition in proton MRI. Magn. Reson. Med. 2009 Sep;62(3):656-64.
3. Hughes P.J, Horn F.C, Collier G.J, et al. Spatial fuzzy c‐means thresholding for semiautomated calculation of percentage lung ventilated volume from hyperpolarized gas and 1H MRI. J. Magn. Reson. Imaging, 2018 47: 640-646.
4. Bakator M, Radosav D. Deep Learning and Medical Diagnosis: A Review of Literature. Multimodal Technologies Interact. 2018; 2:47.
5. Çiçek Ö, Abdulkadir A, Lienkamp S.S et al. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. MICCAI 2016. Lecture Notes in Computer Science, vol 9901. Springer, Cham.
6. Li W, Wang G, Fidon L et al. On the Compactness, Efficiency, and Representation of 3D Convolutional Networks: Brain Parcellation as a Pretext Task. IPMI 2017. Lecture Notes in Computer Science, vol 10265. Springer, Cham.
7. Biancardi A.M, Wild J.M. New Disagreement Metrics Incorporating Spatial Detail – Applications to Lung Imaging. MIUA 2017. Communications in Computer and Information Science, vol 723. 2017. Springer, Cham.
Figures