3186
High-resolution head and C-spine MRI using a Zero-TE (ZTE) sequence at 3T
Aiming Lu1, Carrie M Carr1, Norbert G Campeau1, Steven A Messina1, and David F Kallmes1
1Radiology, Mayo Clinic, Rochester, MN, United States
1Radiology, Mayo Clinic, Rochester, MN, United States
Synopsis
Zero TE (ZTE) sequences have been demonstrated to be capable of achieving CT-like images of the skull and C-spine. To obtain high-resolution images of the skull and c-spine, two separate acquisitions are often performed. Although in general cortical bone, soft tissue and air could be differentiated based on the signal intensity, fatty tissues and connective tissues can be problematic due to the chemical shift artifacts and/or having similar signals to that of cortical bones. This work demonstrated that with optimized protocol and advanced post-processing, ZTE MRI allows for high quality skull and C-spine images in a single acquisition.
Introduction
“Bright-bone” MRI using ultrashort echo time (UTE) and Zero TE (ZTE) sequences have been demonstrated to be capable of achieving CT-like images of the cortical bones (1,2). To obtain high-resolution skull and C-spine MRI images, two separate acquisitions are usually performed. Although in general cortical bones, soft tissue and air could be differentiated based on their signal intensity, fatty tissues and connective tissues can be problematic for visualizing C-spine due to the chemical shift artifacts and/or having similar T2s to that of cortical bones. This work demonstrates that with optimized acquisition protocol and advanced post-processing and the intrinsic large coverage of ZTE MRI, high quality skull and C-spine images could be obtained in a single acquisition at 3T.Materials and Methods
Imaging experiments were performed on a PRISMA 3.0T scanner (Siemens Healthineers, Erlangen, Germany) using a customized Zero TE sequence. A 64-channel head-neck coil was used for imaging. IRB approval was obtained for all healthy human studies. Acquisition parameters included field-of-view(FOV)/pixel bandwidth (BW)/flip angle/TR = 22cm / 343Hz/pixel / 2°/ 1.6 ms. The acquisition matrix was 416 × 416 ×416, resulting in a 0.54 mm isotropic nominal spatial resolution. The acquisition time was 5’54’’. Data were oversampled by a factor of two in the readout direction, which effectively doubled the supported FOV. Relatively high receiver bandwidth was used here to minimize chemical shift artifacts.Image reconstruction was carried out offline. The k-space data were first re-sampled onto Cartesian grids. Fast Fourier transform was then used to generate the magnitude images. The reconstructed matrix size was 416× 416×560 without zero-padding, resulted in a reconstructed FOV of 22cm × 22cm × 30cm. The reconstructed images were processed in Matlab (Mathworks, Natick, MA) to obtain the “bright-bone” images. Due to significant signal inhomogeneity across the large FOV, signal inhomogeneity correction was critical. This was achieved by first normalizing the signal intensity to the mean soft tissue signal in the slice on a slice-by-slice basis, which was then followed by a multi-resolution ROI based intensity correction (1). After correction, the signal intensity in the resultant images was then inverted and the logarithm was taken. Air was removed using a mask generated based on the histogram of the signal intensity. A fast and flexible denoising convolutional neural network (FFDNet) with a tunable noise level map was used to reduce noise, smooth soft tissue contrasts while reserve the details of the cortical bone (3).Results and Discussion
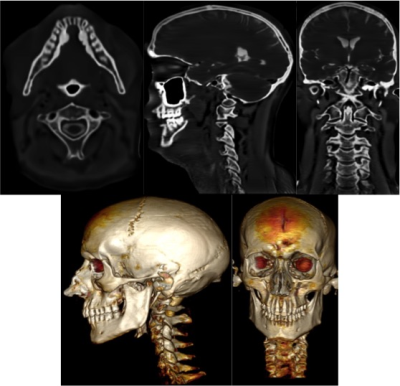
Representative “bright-bone” images of the skull and C-spine of a healthy volunteer are shown in Fig.1. As can be seen, with the proposed data acquisition and image processing, good details of short T2 tissues such as the skull, teeth and the cervical spine can be appreciated in the reformatted sections in different planes. With a proper noise level input, the FFDNet algorithm helped to minimize the soft tissue contrast while still reserving the edge information of the structures of interest. The high signal to noise ratio (SNR) achieved allowed volume-rendered images of the skull and C-spine to be generated.The cerebrospinal fluid (CSF) also appears bright in the reformatted images. This didn't obstruct the visualization of the skull and C-spine here. However, it could be avoided by using a lower flip angle (e.g., 1°) at the cost of decreasing SNR if needed.