3174
Optimized water T2 mapping from multi-echo spin-echo acquisitions with an external fat fraction constraint1Radiological Physics, Department of Radiology, University Hospital of Basel, Basel, Switzerland, 2Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland, 3Neurological Institute Foundation Casimiro Mondino (IRCCS), Pavia, Italy, 4Translational Imaging in Neurology, Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland, 5Department of Neurology, University Hospital of Basel, Basel, Switzerland, 6UMR7357 Laboratoire des sciences de l'Ingénieur, de l'Informatique et de l'Imagerie (ICube), Strasbourg, France, 7Institute of Neurology, A. Gemelli University Hospital Foundation, Catholic University of the Sacred Heart, Rome, Italy, 8Institute of Neurology, Catholic University of the Sacred Heart, Rome, Italy, 9Department of Brain and Behavioral Sciences, University of Pavia, Pavia, Italy, 10Western New York Stem Cell Culture and Analysis Center, Jacobs School of Medicine and Biomedical Sciences, University of Buffalo, Buffalo, NY, United States, 11Fondazione Don Carlo Gnocchi Onlus (IRCCS), Milan, Italy
Synopsis
The measurement of the T2 of the water component is an important marker of neuromuscular diseases. Strategies exist to estimate it from conventional multi-echo spin-echo acquisitions, taking into account fat infiltration and B1 inhomogeneity. In this work, we present an open software that is able to efficiently perform this kind of fitting, and additionally to incorporate the information from an external fat fraction acquisition, for increased accuracy and reduced scan time.
Introduction
Neuromuscular disorders are a broad class of genetic and acquired diseases of the nervous or muscular systems, generally causing different degrees of motion impairment in the affected patients.Imaging has become a valuable tool in the assessment of these diseases, and, specifically, quantitative MR imaging provides robust biomarkers for the monitoring of disease progression. Specifically, T2 of the water component of the muscle tissue and the percentage of fat infiltration are among the most important parameters for the evaluation of edema and degeneration1.Two-component extended phase graph (EPG) fitting of a conventional multi-slice multi-echo spin-echo (MESE) acquisition jointly estimates the fat fraction (FF) and compensates for B1 field inhomogeneity2,3 . While this method has many advantages, including the usage of a standard, widely available MRI sequence, one drawback is the necessity of using a large number of echoes (17 in the proposed implementation) to capture the long T2 of the fat.
In this work, we propose using the information from an external FF acquisition as prior knowledge in the fitting, and we demonstrate that the number of echoes required for accurate fitting can be reduced, thus enabling a better volumetric coverage of the organ of interest in the same scan time. The postprocessing software is released as open-source and is optimized to use a general-purpose graphics processing unit (GPGPU) for fast postprocessing and map creation.
Methods
A stand-alone, command-line application was developed in Python, using standard mathematical extension libraries (NumPy, SciPy) and, for hardware acceleration, the pycuda extension for interfacing with general-purpose graphical processing units (GPGPU, GeForce 1060, NVIDIA corporation).The core of the application consisted of a fast, GPU-accelerated implementation of signal simulation based on the extended phase graph concept4. A Carr-Purcell-Meiboom-Gill (CPMG) simulation was performed, adapting the timing and the number of echoes to the actual sequence parameters. Signals corresponding to multiple combinations of water T2 (wT2), FF, and B1 inhomogeneity factors were simulated and used to build a dictionary, containing a total of 121’200 parameter combinations. The time course of each voxel was then compared to each dictionary entry by using the correlation metric.
When an externally derived FF map was also provided, it was automatically coregistered with each slice of the original spin-echo data. The information was then taken into account by restricting the search of the matching to the given FF for each voxel.
The code was released under a free software license (GNU General Public License v3) at the website: https://www.github.com/fsantini/MyoQMRI.
A conventional 2D multi-slice multi-echo spin-echo (MESE) (17 echoes, 7 slices, TR 4100ms, first TE and echo spacing 10.9ms, resolution 1.2x1.2x10mm3) and a 3D multi-echo gradient-echo (MEGE) (6 echoes, TR 35ms, first TE/echo spacing 1.7/1.5ms, flip angle 7°, resolution 1.0x1.0x5.0 mm³) were acquired with a 3T scanner on the thighs of 5 subjects with diagnosed facioscapulohumeral muscular dystrophy (FSHD), and 4 subjects without a history of neuromuscular diseases. The acquisitions were performed in accordance with the regulations of the local ethics and informed consent was obtained from the participants. FF maps were obtained with FattyRiot5.
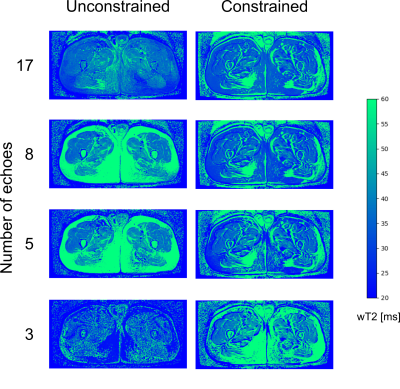
WT2, FF, and B1 maps derived from 17, 8, 5, and 3 echoes with and without the constraint of the external fat map were produced. Average wT2 values inside principal muscle groups and correlation with the FF values were calculated.
Results
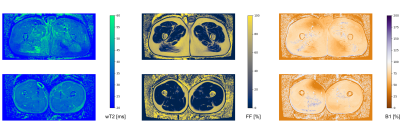
The software fitted the MESE datasets with EPG dictionary matching in an average time of 2 seconds/slice, plus 12 seconds per dataset for the generation of the dictionary.Exemplary outputs from an unconstrained 17-echo reconstruction for a patient and a volunteer are shown in Fig. 1. No noticeable artifacts are visible in the wT2 and FF maps, with only minor inconsistencies in the B1 maps. WT2 maps with 17, 8, 5, and 3 echoes with and without FF constraint are shown in figure 2. Maps with a reduced number of echoes showed lower homogeneity in the unconstrained fitting, whereas the constrained fitting had a more homogeneous appearance but some misregistration artifacts at the edge of the muscles.
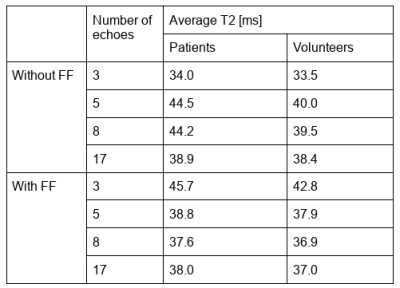
Average wT2 values for patients and volunteers are presented in table 1.
Fitting wT2 with 5 or 8 echoes showed a highly significant correlation with FF (r=0.86 and 0.88, p<0.001), but the correlation was not significant with 17 echoes (p = 0.13), or for the constrained fitting with 5, 8, or 17 echoes (p = 0.25, 0.40, and 0.68, respectively).
Discussion and conclusion
The presented software enables a fast, reproducible reconstruction of wT2 maps with FF estimation from conventional multi-echo spin-echo acquisitions. When an unconstrained reconstruction is used, a minimum number of 17 echoes for the reconstruction is required, otherwise, the T2 values are influenced by the fat infiltration inside the muscle. The usage of an external fat map to constrain the reconstruction allowed the usage of a reduced number of echoes with a minimal penalty in the accuracy of the method.Acknowledgements
This work was supported by the Swiss National Science Foundation (SNSF) grant n° 172876 and by the Italian Ministry of Health (RC 2017-2019, RC 2020 and RF-2016-02362914).References
1. Carlier PG, Marty B, Scheidegger O, et al. Skeletal Muscle Quantitative Nuclear Magnetic Resonance Imaging and Spectroscopy as an Outcome Measure for Clinical Trials. J Neuromuscul Dis 2016;3:1–28.
2. Marty B, Baudin P-Y, Reyngoudt H, et al. Simultaneous muscle water T2 and fat fraction mapping using transverse relaxometry with stimulated echo compensation. NMR Biomed 2016;29:431–443.
3. Keene KR, Beenakker J-WM, Hooijmans MT, et al. T2 relaxation-time mapping in healthy and diseased skeletal muscle using extended phase graph algorithms. Magn. Reson. Med. 2020;84:2656–2670 doi: 10.1002/mrm.28290.
4. Weigel M. Extended phase graphs: Dephasing, RF pulses, and echoes - pure and simple. J. Magn. Reson. Imaging 2015;41:266–295 doi: 10.1002/jmri.24619.
5. Welch EB, Smith DS, Avison MJ, Berglund J, Kullberg J, Ahlström H. Fattyriot - Final Winning Entry Of The 2012 Ismrm Challenge On Water-Fat Reconstruction. Zenodo; 2015. doi: 10.5281/ZENODO.16741.
Figures