3168
Establishing a computational approach to investigate the biophysical basis of relaxivity contrast imaging in the context of muscle degeneration1Imaging Research, Barrow Neurological Institute rrow Neurological Institute, Phoenix, AZ, United States, 2Imaging Research, Barrow Neurological Institute, Phoenix, AZ, United States
Synopsis
The relaxivity contrast imaging parameter, TRATE (tissue transverse relaxivity at tracer equilibrium), has been shown to decrease during ALS induced myofiber degradation, due to T1 and T2* effects that occur when the contrast agent distributes around myofibers. The goal of this study is to expand a validated computational method to investigate the biophysical basis of TRATE in muscles of ALS patients and to optimize RCI acquisition parameters.
Introduction
Loss of spinal and cortical motor neurons are the hallmark of Amyotrophic Lateral Sclerosis (ALS), resulting in progressive muscle atrophy. Due to the clinical heterogeneity of disease presentation and progression robust biomarkers that can be used as surrogates of progression to provide faster and improved decision-making during clinical trials are of interest. Previously published simulations and in vivo validation in brain tumors [1], demonstrated the potential of interrogating cellular features using a susceptibility contrast agent-based imaging approach termed Magnetic Resonance Cytography (MRC) relaxivity contrast imaging (RCI). In particular, the MRC-RCI-derived biomarker, “tissue transverse relaxivity at tracer equilibrium” (TRATE) was found to be highly sensitive to cellular size, density, geometry, and heterogeneity. Our goal is to establish a computational approach to demonstrate the feasibility of TRATE to detect myofiber degeneration.Methods
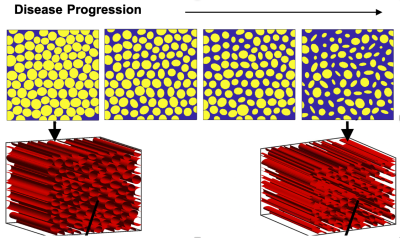
A validated computational strategy termed the finite perturber finite difference method is used to compute MRI signals for 3D tissue structures, detailed description were provided previously [2,3]. Simulation input 3D tissue structures that mimic the known histopathologic progression of ALS-induced muscle degeneration modeled using packed elliptical cylinders were used (Figure 1). Each myofiber property can be individually adjusted (e.g. diameter, aspect ratio, angle, etc) in order to systematically characterize their influence on MRC and also capture the heterogeneity observed during disease progression. Additional input parameters include CA transfer coefficient (Ktrans), water diffusion coefficient and pre-contrast T1, were distinctly chosen to represent values observed in muscle. A multi-echo GRE pulse sequence is modeled to quantify the T2* and T1 changes (ΔR2*(t) and ΔR1(t)) that occur after the injection of a contrast agent (CA). TRATE values are then computed using ΔR2*/Ct for the last few averaged time points. Similar to DCE-MRI, the tissue CA concentration (Ct) is computed using ΔR1/r1, where r1 is the CA’s (known) T1 relaxivity.Results and Discussion
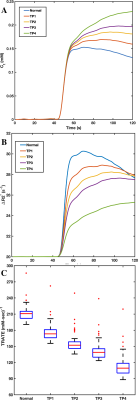
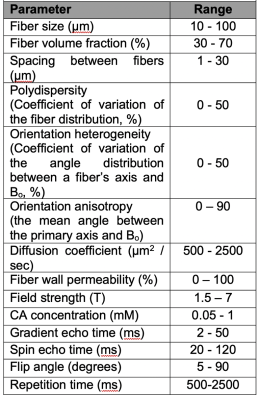
Figure 1 illustrates example muscle tissue structure used to characterize RCI in silico, as a function of ALS progression achieved by inducing loss of myofiber diameter. Top row shows a 2D section of the 3D structure below. Figure 2A-C shows mean Ct, mean ΔR2* and computed TRATE values across all simulated muscle tissue structures at various disease manifestation time points (TP). These preliminary results show TRATE values decrease as myofiber degrade. The decrease in TRATE most likely originates from the decrease in myofiber volume fraction consistent with previous results [1]. As with any computational model, challenges can arise when trying to accurately recapitulate the in vivo tissue properties. A critical element of our simulations is the structure used to mimic individual myofibers as wells size, relative average orientation with respect of the main magnetic field, water diffusion coefficient. The complex relationship between muscle microstructure and diffusion profile and its influence on the derived RCI parameter can also be adjusted by using restricted and unrestricted diffusion models. Constrained diffusion of the contrast agent around fibers within the voxel could also result heterogeneous susceptibility distribution and can be modeled using a similar approach as water diffusion. Table 1 summarizes the structural, physiological, and pulse sequence parameter currently under investigation as part of the optimization process.Conclusion
Results of this study provide confirmation of our prior in vivo results that TRATE can longitudinally differentiate myofiber atrophy in an ALS model. To establish TRATE as a potential biomarker of disease progression and therapy response in patients with ALS we are refining the proposed computational approach in order to optimize acquisition techniques and investigate its sensitivity across a wide range of biophysical parameters.Acknowledgements
NCI R01 CA158079References
1. Semmineh, N. B., Xu, J., Skinner, J. T., Xie, J., Li, H., Ayers, G., & Quarles, C. C. (2015). Assessing tumor cytoarchitecture using multiecho DSC- MRI derived measures of the transverse relaxivity at tracer equilibrium (TRATE). Magnetic Resonance in Medicine, 74(3), 772–784.
2. Semmineh NB, Stokes AM, Bell LC, Boxerman JL, Quarles CC. A population based digital reference object (DRO) for optimizing dynamic susceptibility contrast (DSC) MRI methods for clinical trials. Tomography n.d.
3. Semmineh NB, Xu J, Boxerman JL, Delaney GW, Cleary PW, Gore JC, et al. An efficient computational approach to characterize DSC-MRI signals arising from three-dimensional heterogeneous tissue structures. PLoS One 2014;9. doi:10.1371/journal.pone.0084764.
Figures