3141
Validation of group cohesive parcellation of rsfMRI1Cleveland Clinic, Cleveland, OH, United States
Synopsis
Single-subject cohesive parcellation were done
for nine healthy subjects’ rsfMRI data.
Based on motion quality, the best data for six subjects was used for
group cohesive parcellation. The parcellation was in a leave one out cross
validation. DICE scores were similar to intra-subject cross-validation and
significantly better than random parcellation.
Introduction
Parcel cohesion is a measure of how well the mean parcel time course correlates with its underlying members voxels. Traditional connectivity-based parcellation yields poor parcel cohesion (Yeo 2011, Choi 2012). Previously, single subject cohesive parcellation using spatially-constrained agglomeration hierarchical clustering was shown to produce parcels with excellent cohesion (Nemani 2020). Here, we extend single subject cohesive parcellation to the group level and evaluate its reproducibility in a leave one out cross-validation scheme.Method
Data CollectionNine healthy subjects without neurological disease were recruited and scanned as controls for NMSS study at two time points (2 years apart). There were one T1 MPRAGE scan and two functional connectivity MRI each time. Task-free fMRI was performed with a Prisma 3T scanner (Siemens, Erlangen, Germany) using a 20-channel receive-only head coil and a bite bar to restrict head motion.
Scan 1: Whole-brain T1 MPRAGE: 176 axial sections; thickness, 0.940 mm; FOV 256*256 mm; TI/TE/TR/flip angle, 900/1.71/1900 ms/8°; matrix, 256*176; band width, 62 kHz.
Scan 2: Whole-brain resting-state fcMRI study, with eyes closed: 128 repetitions of 31 4-mm-thick axial sections (no gap); matrix, 128*128; in-plane resolution, 2*2 mm; TE/TR/flip angle, 29/2800 ms/80°; FOV, 256*256 mm2; band width, 1954 Hz/pixel.
Data Analysis
Images of functional connectivity data were brain extracted with FSL, motion corrected with SLOMOCO and corrected for physiological nuisances with PESTICA (Beall, 2007; Beall, 2014). Anatomical data were warped to rsfMRI data with FreeSurfer and ANTS. All images are warped to the same template (one subject’s fcMRI) with the isotropic resolution of 2*2*2 mm. Across all 9 subjects, there were 36 total rsfMRI datasets collected. The 36 scans were analyzed and three subjects were rejected due to motion. Single subject cohesive parcellation was performed for each of the four scans from these six subjects and evaluated using intra-subject cross-validation of DICE scores (Nemani 2020).
The best scan from each subject was used for group cohesive parcellation. Briefly, group cohesive parcellation extends single subject parcellation by pooling the mean parcel cohesion at each step of agglomeration in a parallel hierarchical clustering framework. The two clusters that produced the highest pooled cohesion using a robust 10th percentile threshold were selected for merging at each step until a full group agglomerative tree was generated. This tree was cut at a cohesion threshold of 0.5 to produce final group parcels. Reproducibility was evaluated using DICE scores in a leave one out cross-validation.
Results
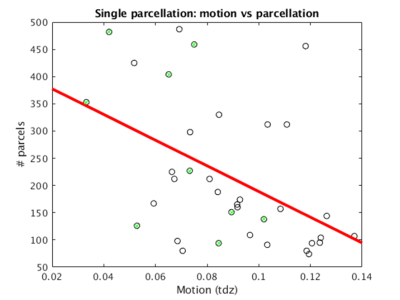
Over all 36 scans across the nine healthy subjects, the mode of tdz motion was 0.089±0.026 (Beall 2014). Three subjects (4, 5, and 6) were rejected due to an overall motion quality threshold of 0.080. The optimal scan for each subject (1, 2, 3, 7, 8, and 9) was selected. The mode of tdz motion for the selected six scans was 0.057±0.017.Figure 1 shows number of parcels vs motion for all 36 single subject parcellations. The optimal scans for each subject are highlighted in green. The six optimal scans (tdz motion < 0.080) were used for group cohesive parcellation.
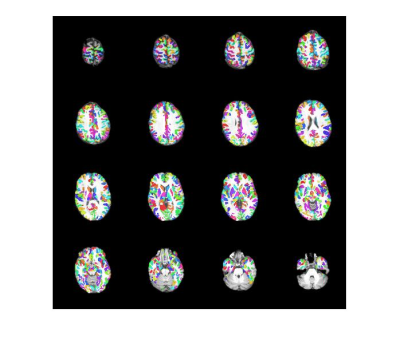
Figure 2 shows the group cohesive parcellation for the group of the six subjects. All parcels had a cohesion greater than 0.5 as designed.
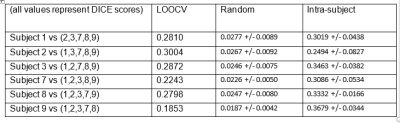
Table 1 shows DICE scores for leave one out cross-validation (LOOCV), random parcellation (null) and intra-subject cross-validation (Intra-subject). Random parcellation was generated using 100 random circular shifts of the cohesive parcels to preserve underlying structure.
Discussion
There is a clear trend that the more motion there was in the data, the less parcels extracted (Figure 1, red line), which can be explained by the fact that gross head motion by nature introduces spatiotemporal correlations into rsfMRI data. Motion-based correlations reduces the number of parcels produced, which can potentially be a valuable independent tool to evaluate the quality of the underlying data.Based on leave one out cross-validation using DICE analysis, group cohesive parcellation was significantly more reproducible than random labeling, and had similar reproducibility to intra-subject cohesive parcellation (Table 1).
Conclusion
Cohesive parcellation is based on ensuring that parcel exemplar time series best represent their underlying parcel members. Single subject cohesive parcellation was extended to the group level and shown to have reasonable reproducibility while still preserving excellent cohesion at the group level. This enables optimal downstream analyses at both the single subject and group levels with parcels that have a straightforward interpretation.Acknowledgements
This work was supported by the Imaging Institute, Cleveland Clinic.
Authors acknowledge technical support by Siemens Medical Solutions.
References
Yeo, B.T., et al (2011). The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol 106, 1125-1165.
Choi, E.Y., et al (2012). The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol 108, 2242-2263.
Nemani, A., Lowe, M.J. (2020). Cohesive parcellation of rsfMRI using constrained hierarchical clustering. ISMRM 2020, poster # 3960.
Beall, E.B., Lowe, M.J. (2007). Isolating physiologic noise sources with independently determined spatial measures. Neuroimage 37, 1286-1300.
Beall, E.B., Lowe, M.J. (2014). SimPACE: generating simulated motion corrupted BOLD data with synthetic-navigated acquisition for the development and evaluation of SLOMOCO: a new, highly effective slicewise motion correction. Neuroimage 101, 21-34.