3139
Locating seed automatically in posterior cingulate cortex for resting state fMRI data analysis by using unsupervised machine learning1Imaging Institute, Cleveland Clinic, Cleveland, OH, United States
Synopsis
We developed an automatic pipeline to generate seed clusters and corresponding connectivity maps for rs-fMRI data analysis by using unsupervised machine learning method. It only needed manual participation in the very end to review the candidate seed cluster locations and connectivity maps to make decision. Seeds in our pipeline were determined functionally within large pre-defined ROI which could be derived by using automatic brain segmentation tools like FreeSurfer or image registration. Successful application of the pipeline to locate seeds in PCC of control subjects and patients will be presented in this abstract.
Introduction
When using seed-based methods to analyze resting state fMRI (rs-fMRI) data, the selection of the seed has large influence on the results. In previous study, we presented a method to automatically produce seed locations from voxel clusters generated by self-organizing map (SOM) whose input was feature vectors formed by combining anatomical and rs-fMRI data[1]. In this study, we tested seed generation in posterior cingulate cortex(PCC) of control subjects and patients by using this method against a manual method using “instacorr” tool in AFNI[2].Methods
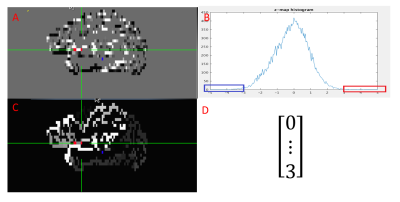
Data acquisition: Twelve subjects consisting six healthy controls and six patients were scanned in an IRB-approved protocol at 3T Siemens scanner (Erlangen, Germany) using a bitebar to reduce head motion, in a 12-ch receive head coil. Scans included T1-MPRAGE (voxel size=1x1x1.2mm, matrix size=256x256x120, TE/TR/TI=1.75/1900/900ms), and rs-fMRI(voxel size=2x2x4mm, matrix size=128x128x31, TR/TE/FA=2800/29/80, 132volumes).Data processing: Each rs-fMRI dataset was motion-corrected, low-pass filtered and spatially filtered. T1 image was parcellated into ROIs by using FreeSurfer[3]. The ROIs were registered to rs-fMRI image space by using “align_epi_anat.py” tool in AFNI. The ROI covering the left/right PCC was used as seed searching region for automatic seed generation method (Left side and right side are processed separately). From rs-fMRI data, the global connectivity between each voxel in the seed searching ROI and all other brain cortex voxels were computed and then the connectivity distribution was fitted into a Gaussian distribution[4]. The feature vector was formed by counting the number of voxels whose connectivity value was outside three standard deviation , in the parcellated cortex ROIs. The above feature vector forming step is shown in figure 1. Then the feature vectors of all the voxels in the searching region was feed into a size 4x4 SOM classifier in Matlab. Seed clusters were derived from the voxel clusters corresponding to the classified feature vector clusters. The connectivity map of a seed cluster was the average of the connectivity maps computed for all the voxels in the seed cluster.
Using each subject’s rs-fMRI data, seeds in PCC were also manually located by experts through the “instacorr” method in AFNI.
Seed comparison: Seeds acquired through the automatic method were compared to those picked through the manual method.
Results
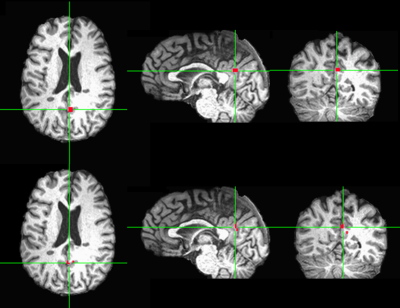
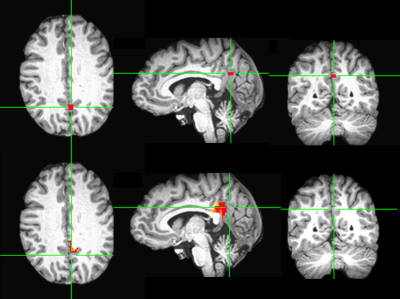
In four out of six control subjects, there was one automatically generated seed overlapping with manually picked seed. In the two unmatched control subject cases, the manually located seed was outside the seed searching ROI of the automatic method. In three out of six patients, there is one automatically generated seed overlapping with manually picked seed. The reason for three unmatched patient cases was the same as the unmatched control cases. One matched patient case is shown in figure 2 and one unmatched patient case is shown in figure 3.Discussion and Conclusion
The automatically seed generation method could generate seed in PCC to match the manually picked seed as long as the seed searching ROI included the manually picked seed.The manually produced seed usually takes the shape of a cubic. In contrast, automatically generated seed follows the anatomical structure as shown in the figure 2.
Propagation of a template ROI to individual subjects through image registration can be an alternative way to establish seed searching ROI for automatic method. We will test it out in our future study and we hope it can help to overcome the unmatched cases.
Acknowledgements
This work was supported by the Imaging Institute, Cleveland Clinic. Authors acknowledge technical support by Siemens Medical Solutions.References
[1]Li M, et al. Automatic seed selection for resting state fMRI data analysis by using machine learning, ISMRM 27th Annual Meeting & Exhibition, Montreal, Canada, May 2019
[2] Cox RW. AFNI: What a long strange trip it’s been, NeuroImage. 2012; 62(2):743-747.
[3] Jenkinson M, et al. FSL, NeuroImage.2012; 62(2):782-790.
[4] Lowe MJ, et al. Functional Connectivity in Single and Multislice Echoplanar Imaging Using Resting-State Fluctuations, NeuroImage. 1998; 7(1):119-132.
Figures