3063
Faster Bloch simulations and MR-STAT reconstructions on GPU using the Julia programming language1Computational Imaging Group for MR Diagnostics and Therapy, Center for Image Sciences, University Medical Center Utrecht, Utrecht, Netherlands, 2Department of Radiology, Division of Imaging and Oncology, University Medical Center Utrecht, Utrecht, Netherlands
Synopsis
MR-STAT is a multi-parametric quantitative MR framework with computationally demanding reconstructions. In this work we implemented the reconstruction algorithm on the GPU using the Julia programming language, a language that allows for quick prototyping without sacrificing on performance. We demonstrate superior runtime-performance of the GPU implementation as compared to previously proposed implementations running on a cluster of CPU's. With the proposed implementation, high-resolution in-vivo parameter maps can be reconstructed in approximately two minutes. The proposed implementation can also be used to rapidly generate MR Fingerprinting dictionaries and is shown to outperform the native CUDA implementation from SnapMRF.
Introduction
MR-STAT is a recently proposed quantitative MR framework for obtaining multiple parameter maps from a single short scan.1 The parameter maps $$$\boldsymbol{\alpha}$$$ (e.g. $$$T_1,T_2$$$ and $$$\rho$$$ maps) are reconstructed from measured time-domain data $$$\mathbf{d}$$$ by numerically solving $$\text{argmin}_\boldsymbol{\alpha} \| \mathbf{d} - \mathbf{s}(\boldsymbol{\alpha})\|_2^2,$$where $$$\mathbf{s}$$$ is the sequence-dependent volumetric signal model. Solving this large-scale, non-linear optimization problem is a computationally demanding task: in previous works reconstruction times of a few hours per slice on a CPU cluster were reported.2 Algorithmic acceleration techniques have been proposed to reduce reconstruction times, but so far these methods rely on the assumption of Cartesian acquisitions.3,4 In the current work we have used the Julia programming language5 to adapt the MR-STAT reconstruction algorithm to GPU architectures. We demonstrate superior runtime-performance of the GPU implementation as compared to previously proposed reconstructions on a cluster of CPU's for both Cartesian and radial acquisitions. In addition, the GPU implementation is also used to rapidly generate dictionaries for MR Fingerprinting6 and the runtimes are shown to be approximately five times faster compared to a native CUDA implementation from snapMRF.7Methods
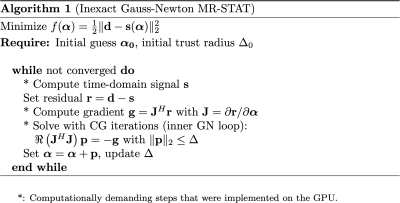
All computationally demanding steps of the previously proposed Gauss-Newton MR-STAT reconstruction algorithm3 (see Figure 1) were implemented using the Julia (v.1.5.3) programming language. This is a relatively new programming language that offers the flexibility and ease-of-use of dynamically-typed, interpreted languages while - through novel language design in combination with a “Just-in-time” compiler - giving performance similar to that of statically-typed, compiled languages. The CUDA.jl package8 was used to write kernel functions in Julia that can be executed in parallel exploiting CUDA hardware (as well as on CPU through the use of KernelAbstractions.jl9), allowing for highly flexible yet performant code.To demonstrate the performance of the GPU-based reconstruction, data from a single brain slice was obtained from healthy volunteer using a 3T clinical MR system (Ingenia, Philips Healthcare). For the acquisition, a gradient-spoiled pulse sequence with variable flip angle train, starting off with an adiabatic inversion pulse was used. The TR, TE and inversion time were 8.9 ms, 4.7 ms and 10 ms, respectively. The in-plane resolution was 1 mm2 and the slice thickness 5 mm. We used both Cartesian and radial readout trajectories to acquire 1120 readouts resulting in a scan time of 9.9 s.
Slice profile effects were modelled using the the small tip-angle approximation. For both the Cartesian and radial acquisition, 6 iterations of the Gauss-Newton method were sufficient for convergence. The GPU reconstructions were performed on a Quadro RTX 6000 GPU card. Runtimes were compared to runtimes reported in previous CPU-based works.2,3,4,10
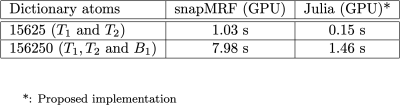
Part of the MR-STAT reconstruction code is a standalone Bloch (EPG) simulator that could also be used to generate MRF dictionaries. To demonstrate the performance of our Julia-based GPU implementation, we generated two dictionaries and compared the runtimes to the recently proposed snapMRF7 , which is based on a low-level CUDA implementation and is optimized for performance. One dictionary contains $$$T_1$$$ and $$$T_2$$$ dimensions only (15625 atoms). The second dictionary also includes $$$B_1^+$$$ (156250 atoms).
Results
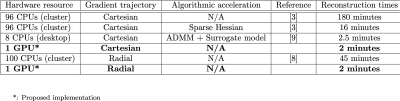
In Fig. 2 the reconstruction times for the Cartesian and radial MR-STAT reconstructions on the GPU are shown. It can be seen that a single GPU card gives better runtime-performance than 96 CPUs on a computing cluster. The GPU implementation also slightly outperforms the ADMM-based MR-STAT reconstruction method proposed in [3] that runs on a regular desktop computer. That method, however, relies on Cartesian acquisitions and cannot be used for e.g. radial acquisitions. The current GPU implementation on the other hand also significantly reduces the reconstruction times for radial acquisitions.In Fig. 3, the times needed to simulate the MRF dictionaries are reported. It can be seen that our Julia-based GPU implementation is approximately five times faster than SnapMRF. At the same time, because the code is written in a high-level language, adapting the underlying model by making changes to the pulse sequence or adding new tissue parameters is relatively easy.
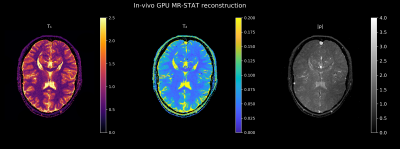
In Fig. 4 we display the high-resolution $$$T_1, T_2$$$ and $$$\rho$$$ maps that were reconstructed from the Cartesian in-vivo dataset. The reconstruction time was two minutes.
Discussion and Conclusion
We have demonstrated that MR-STAT reconstructions are suitable for execution on GPUs and that the performance is improved compared to using a cluster of CPUs. By using the Julia language and its rich package ecosystem, we were able to write the reconstruction code (i.e. CUDA-kernels) in a high-level manner without sacrificing on performance. This is desired in a rapidly evolving and computationally demanding field such as qMRI, where fast runtimes are important and at the same time flexibility is required in sequence design and/or the number of distinct tissue parameters to be included in the model.Even though MR-STAT reconstruction times have been reduced compared to the CPU-based implementations, the reconstruction times are still long for clinical purposes. Further research is dedicated towards combining the GPU-based implementation with previously proposed algorithmic improvements3,4 to the reconstruction algorithm as well as deep-learning based acceleration methods4,11 with the aim of reaching clinically acceptable reconstruction times for MR-STAT.
Acknowledgements
This research is funded by the Netherlands Organisation for Scientific Research, domain Applied and Engineering Sciences, Grant #14125.References
[1] Sbrizzi, Alessandro, et al. "Fast quantitative MRI as a nonlinear tomography problem." Magnetic resonance imaging 46 (2018): 56-63.
[2] van der Heide, Oscar, et al. "High‐resolution in vivo MR‐STAT using a matrix‐free and parallelized reconstruction algorithm." NMR in Biomedicine 33.4 (2020): e4251.
[3] van der Heide, Oscar, Alessandro Sbrizzi, and Cornelis AT van den Berg. "Accelerated MR-STAT reconstructions using sparse Hessian approximations." IEEE Transactions on Medical Imaging 39.11 (2020): 3737-3748.
[4] Liu et al. “Accelerated MR-STAT Algorithm: Achieving 10-minute High-Resolution Reconstructions on a Desktop PC”, ISMRM 2020, p3477.
[5] Bezanson, Jeff, et al. "Julia: A fresh approach to numerical computing." SIAM review 59.1 (2017): 65-98.
[6] Ma, Dan, et al. "Magnetic resonance fingerprinting." Nature 495.7440 (2013): 187-192.
[7] Wang, Dong, Jason Ostenson, and David S. Smith. "snapMRF: GPU-accelerated magnetic resonance fingerprinting dictionary generation and matching using extended phase graphs." Magnetic Resonance Imaging 66 (2020): 248-256.
[8] Besard, Tim, Christophe Foket, and Bjorn De Sutter. "Effective extensible programming: unleashing Julia on GPUs." IEEE Transactions on Parallel and Distributed Systems 30.4 (2018): 827-841.
[9] https://github.com/JuliaGPU/KernelAbstractions.jl, DOI: 10.5281/zenodo.4135830
[10] van der Heide, Oscar et al. “Extension of MR-STAT to non-Cartesian and gradient-spoiled sequences”, ISMRM 2020, p0886.
[11] Liu, Hanna et al., "Fast and Accurate Modeling of Transient-State Gradient-Spoiled Sequences by Recurrent Neural Networks", https://arxiv.org/abs/2008.07440
Figures