3062
BUDA-SAGE with unsupervised denoising enables fast, distortion-free, high-resolution T2, T2*, iron and myelin susceptibility mapping1State Key Laboratory of Modern Optical Instrumentation, College of Optical Science and Engineering, Zhejiang University, Hangzhou, China, 2Department of Radiology, A. A. Martinos Center for Biomedical Imaging, Massachusetts general hospital, Boston, MA, United States, 3Subtle Medical Inc, Menlo Park, CA, United States, 4Laboratory for Imaging Science and Technology (LIST), Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of
Synopsis
We propose BUDA-SAGE, an efficient echo‐planar imaging (EPI) sequence for quantitative mapping. The acquisition includes multiple T2*-, T2’- and T2-weighted contrasts. We alternate the phase-encoding polarities across the shots in this multi-shot navigator-free acquisition to eliminate geometric distortion. An unsupervised Self2Self (S2S) neural network (NN) was utilized to perform denoising after BUDA reconstruction to achieve 1×1×2 mm3 resolution with high SNR. We demonstrate the ability of BUDA-SAGE to provide whole-brain, distortion-free, high-resolution multi-contrast images and quantitative T2, T2* maps in 50 seconds, and separate para- and dia-magnetic susceptibility maps in 140 seconds.
Introduction
Quantitative parameter mapping has demonstrated potential in clinical and neuroscience applications, but its adoption has been hampered by the long acquisition time required to encode multi-contrast images that capture the signal evolution. Employing EPI readout can improve the acquisition speed, but standard 2d-EPI does not lend itself to high resolution imaging due to severe geometric distortion and low SNR.To eliminate distortion, Blip Up- and -Down Acquisition (BUDA)1 collects multiple shots with alternating phase-encoding polarities. It jointly reconstructs these using field map information in the forward model and incorporates Hankel structured low-rank regularization into Hybrid-SENSE2 to eliminate distortion without need for phase navigation.
Unsupervised deep-learning denoising methods have shown great potential in improving image SNR3,4,5, and can complement rapid EPI acquisitions. Self2Self (S2S)5 is a novel network trained on Bernoulli-sampled instances of the input noisy image. With the reduction of the image variance, S2S learns to generate denoised images to improve SNR effectively.
By combining BUDA with S2S, we propose to harness the efficiency of this distortion-free acquisition in high-resolution imaging with high SNR. To obtain quantitative maps rapidly, we incorporate two additional EPI readouts before and after 1800 refocusing pulse in the spin-echo EPI sequence to acquire gradient-, mixed-, and spin-echo images simultaneously in one scan^6. By changing the echo time, additional contrasts can be acquired. Using BUDA+S2S, we demonstrate whole-brain, distortion-free T2, T2*, para- and dia-magnetic susceptibility maps at 1×1×2 mm3 resolution in 140s.
Methods
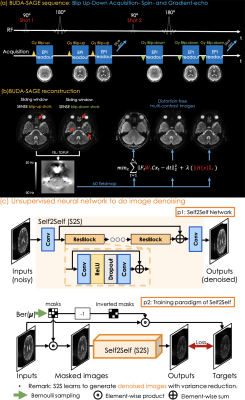
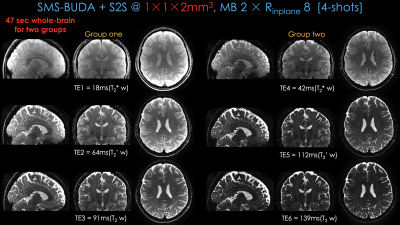
Fig1 (a) shows the proposed sequence where two additional EPI readouts, with gradient and mixed gradient-spin echo contrasts. Multiple shots are sampled with opposite phase-encoding polarities^1. We extend the sequence to simultaneous multi-slice (SMS) encoding7 and quantitative MRI. Multiple scans can provide additional contrasts by changing the TEs of the sequence.Acquisition: To obtain quantitative T2, T2* maps, 1×1×2 mm3 resolution BUDA-SAGE data were acquired at Rinplane=8, SMS=2 with 8-shots using FOV = 220×220×120mm3. We changed the TEs of the sequence and perform three separate scans to acquire three groups of data with [TEgre, TEmixed, TEse]=[18, 64, 91] in group 1, [30, 88, 115] in group 2 and [42, 112, 139] in the last scan at TR=5000 ms. Each group provided three different contrasts. To obtain QSM map and disentangle its para- and dia-magnetic constituents, we collected another three groups of BUDA-SAGE data. The acquisition parameters were same except for FOV = 220×220×128mm3, TR = 5500 ms.
Reconstruction: We first sum the k-space data of spin-echo for the blip-up shots and blip-down shots and perform SENSE to generate interim images as shown in Fig1 (b).We then estimate a field map using topup8 and incorporate it in the joint BUDA reconstruction for the other contrasts:$$\min_{x} \sum_{t=1}^{N_s} \| F_t W_t C x_t - d_t\|_2^2 + \lambda \|H(x)\|_*$$
Where Ft is the undersampled Fourier operator in shot t, Wt is distortion operator in shot t, C is sensitivity map from ESPIRIT9, and dt is the k-space data of each shot. ||H(x)||* is Hankel structured low-rank constraints.
We use all 8-shots data to provide a reference reconstruction. A subset of 4-shot are used to reduce the acquisition time by half.
Denoising: unsupervised NN S2S aims to denoise the reconstructed 4-shot images, and does not require ground-truth clean images for training. Taking noisy images as inputs, S2S employs multiple convolution layers and ResBlocks5 to generate the denoised results.
To train S2S, we use masks m whose elements are iid sampled from a Bernoulli distribution, and we obtain the corresponding inverted masks, 1-m. The mask m is applied on the noisy images to form the input, and the inverted mask is applied on them to generate target images. We minimize the L1-norm loss between the outputs and targets to train the S2S network.
Quantitative mapping: T2 (=1/R2) and T2* (=1/R2*) maps are obtained using Bloch dictionary matching on the reconstructed echoes. QSM is estimated using three gradient echoes from three groups data with NDI10. Using the estimated R2 and R2* maps, we also derive the R2’ information required for source separation QSM reconstruction, which provides us with additional para- and dia-magnetic susceptibility maps11.
Results
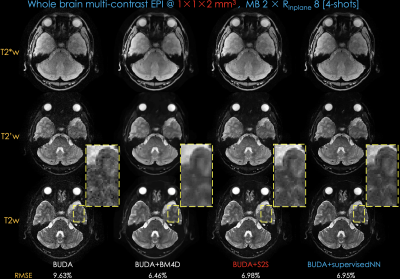
Fig 2 shows distortion-free, high-resolution multi-contrast images including T2*-, T2’- and T2-weighted images using 4-shot data. Compared to BUDA, BUDA+S2S effectively reduced the noise. To validate the effectiveness of S2S, we conducted an experiment with a supervised NN whose structure is the same as S2S. To obtain ground-truth data, we collected 2-averages of 8-shot data with five subjects. The results from BUDA+S2S are comparable with those from BUDA+supervisedNN.Fig 3 shows whole-brain distortion-free six-contrast images from two groups of BUDA+S2S from a 45s scan (20s/group + 5s dummy), plus a 2s calibration scan for coil sensitivities.
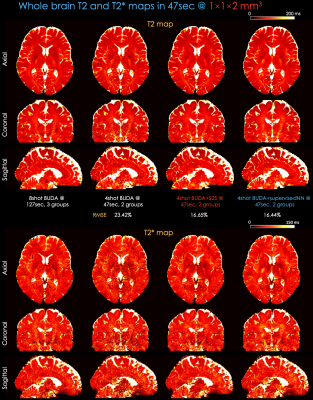
Fig 4 shows whole-brain, distortion-free quantitative T2 and T2* maps at 1×1×2 mm3 resolution in 47s. BUDA+S2S and BUDA+supervisedNN provide comparable maps using 4-shot, 2-groups data with respect to 8-shot, 3-groups data at Rinplane=8, SMS=2.
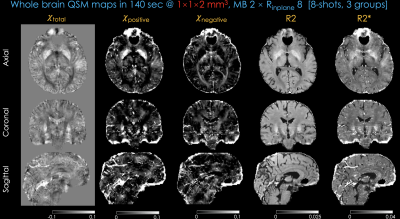
Fig 5 shows whole-brain, distortion-free total, para- and dia-magnetic QSM maps along with R2, R2* maps using 3-groups of 8-shot data.
Conclusion
We demonstrate whole-brain, distortion-free fast and comprehensive quantitative mapping using a synergistic combination of efficient multi-shot SAGE acquisition, BUDA reconstruction and unsupervised S2S denoising.Acknowledgements
This work was supported by research grants NIH R01 EB028797, U01 EB025162, P41 EB030006, U01 EB026996, the NVidia Corporation for computing support, and by the National Natural Science Foundation of China (No: U1809204, 61525106, 61427807, 61701436), by the National Key Technology Research and Development Program of China (No: 2017YFE0104000, 2016YFC1300302), and by Shenzhen Innovation Funding (No: JCYJ20170818164343304, JCYJ20170816172431715).References
1. Bilgic B, Liao C, Manhard MK, et. al, “Robust high-quality multi-shot EPI with low-rank prior and machine learning”, Magn Reson. Med., 2019.
2. Zahneisen B, Aksoy M, Maclaren J, Wuerslin C, Bammer R, “Extended hybrid-space SENSE for EPI: Off-resonance and eddy current corrected joint interleaved blip-up/down reconstruction”, NeuroImage., 2017;153:97-108.
3. Lehtinen J, Munkberg J, et. al, “Noise2Noise: Learning Image Restoration without Clean Data”, ICML, 2018, 80:2965-2974.
4. Krull A, Buchholz TO, Jug F, “Noise2Void - Learning Denoising From Single Noisy Images”, CVPR, 2019, pp. 2129-2137.
5. Quan Y, Chen M,
Pang T, Ji H, “Self2Self With Dropout: Learning Self-Supervised Denoising From
Single Image”, CVPR, 2020, pp. 1890-1898.
6. Zhang Z, Liao
C, Cho J, et. al, “dSAGE enables distortion-free diffusion, spin and gradient
echo imaging in 1 minute”, Magn Reson. Med., 2020.
7. Setsompop K, Gagoski B A, Polimeni J R, et. al, “Blipped‐controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g‐factor penalty”, Magn Reson. Med., 2012;5:1210-1224.
8. Smith, S.M., Jenkinson, M., Woolrich, M.W., Beckmann, C.F., et. al, “Advances in functional and structural MR image analysis and implementation as FSL”, NeuroImage., 2004;23:S208-S219.
9. ecker M, Lai P, Murphy M J, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M, “ESPIRiT-an eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA”, Magn. Reson. Med. 2014;71:990-1001.
10. Polak D, Chatnuntawech I, Yoon J, et. al, “Nonlinear dipole inversion (NDI) enables robust quantitative susceptibility mapping (QSM)”, NMR in Biomedicine., 2020;e4271.
11. Lee J, Nam Y, Choi J et. al, “Separating positive and negative susceptibility sources in QSM.”, Magn Reson. Med., 2017.
Figures