3058
Ultra-high Spatial Resolution Multispectral qMRI with Compressed Sensing Tri-TSE1Boston University, Boston, MA, United States, 2Boston University Medical Center, Boston, MA, United States, 3Center for Translational Neuroimaging, Boston, MA, United States
Synopsis
Purpose: To test the PD-T1-T2 qMRI accuracy of ultra-high spatial resolution compressed sense Tri-TSE. Methods: A healthy male volunteer (35yo) was scanned with an ultra-high spatial resolution compressed sensing Tri-TSE pulse sequence. Maps of PD, T1, and T2 were generated with voxel size of 0.4 x 0.4 x 1.2 mm3. Results: PD, T1, and T2 accuracy are not affected by threefold compressed sensing acceleration. Conclusion: Compressed sensing does not appear to negatively impact MS-qMRI accuracy and opens the door to ultrafast brain MS-qMRI at current clinical spatial resolution or to a new high spatial resolution standard in the clinical context.
Introduction
Increasing the speed of multispectral qMRI (MS-qMRI) acquisitions is of paramount importance for developing clinically practical quantitative protocols, particularly for high spatial resolution applications. Compressed sensing (CS) is a recently developed technique based on the premise that if “the underlying image exhibits transform sparsity, and if k-space undersampling results in incoherent artifacts in that transform domain, then the image can be recovered from randomly undersampled frequency domain data, provided an appropriate nonlinear recovery scheme is used”1. In this work, we tested a commercially available CS implementation in combination with an ultra-high spatial resolution variant of the Tri-TSE pulse sequence to test for MS-qMRI accuracy relative to its non-CS counterpart.Methods
A healthy male volunteer (35yo) was consented with an IRB approved research protocol. We tested CS-Tri-TSE at 3T (Philips, Ingenia Elition X) with a CS factor of 3 to generate self-coregistered maps of the relaxation times (T1 and T2) and proton density (PD) at a native voxel size of 0.4 x 0.4 x 1.2 mm3. Other selected parameters: TRlong=11,142ms, TRshort=550ms, TE1,2=13.75 and 110ms, Ns=130 slices, Scan time=17min. The scanner CS-reconstructed directly acquired dicom images were further processed with MS-qMRI algorithms programmed in Python 3.7 with the Enthought Deployment Manager. Two preparation steps are required and performed with Fiji: 1) manual editing of the segmented intracranial matter (ICM) segment and 2) manual delineation of the cerebellum’s superior aspect. MS-qMRI algorithms were derived as functions of pulse sequence parameters according to the Bloch equation model of the Tri-TSE, which is applicable across all MRI platforms (Eq. 1-3).Eq. 1: $$$T_2=cf_3\frac{TE1_{eff}-TE2_{eff}}{ln\left(\frac{DA_2}{DA_1}\right)}$$$
Eq. 2: $$$PD=\frac{cf_1}{C_{coil}}\cdot\frac{DA_1\exp\left(\frac{TE1_{eff}}{T_2}\right)+DA_2\exp\left(\frac{TE2_{eff}}{T_2}\right)}{1-\exp\left({\frac{-\left(TR_{long}-TSEshot_{DE}\right)}{T_1}}\right)}$$$
Eq. 3: $$$T_1=Root_{hybr}\left\{T_1+\frac{TR_{short}}{ln\left(\frac{1-\left(\frac{DA_3}{DA_1}\right)\left[\left(1-\exp\left(\frac{-TR_{long}}{T_1}\right)\right)-cf_2\exp\left(\frac{-\left(TR_{long}-TSEshot_{DE}\right)}{T_1}\right)\right]}{1+cf_2\exp\left(\frac{TSEshot_{SE}}{T_1}\right)}\right)}\right\}$$$
Results
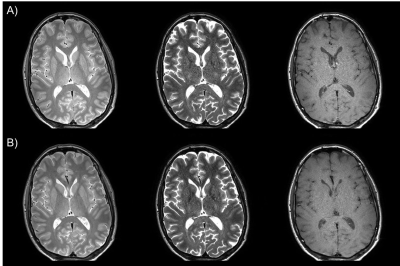
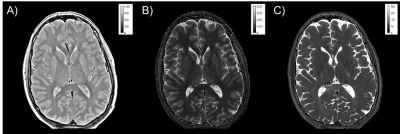
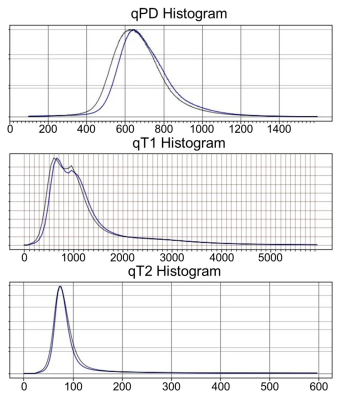
As shown in Fig. 1, the image quality of the CS-Tri-TSE images is nearly indistinguishable from the non-CS counterparts in terms of contrast and SNR. Remarkably, flow ghosting artifacts (not shown) were less severe in the CS-Tri-TSE images, specifically in the posterior fossa. Selected qMRI maps are shown in Fig. 2 with the corresponding pixel value scale bars showing good quantitative agreement with values reported in the literature2 as further confirmed in the whole-brain histograms (Fig. 3). These histograms further show that there is less parameter spread in the CS-maps, possibly indicating less vulnerability to pulsation induced variability.Discussion
We tested a three-fold acceleration factor and found that MS-qMRI accuracy and map quality are not affected. It is likely that further improvements in pulse sequence development and compressed sensing technologies could lead to routine ultra-high spatial resolution in the clinical context.Conclusions
Compressed sensing does not appear to negatively impact MS-qMRI accuracy and opens the door to either, ultrafast brain MS-qMRI at current spatial resolution or to a new high spatial resolution standard in the clinical context. This work could have implications for protocol unification and standardization, and for ultra-high spatial resolution Synthetic-MRI and white matter fibrography.Acknowledgements
No acknowledgement found.References
1. Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magnetic Resonance in Medicine 2007;58(6):1182-1195.
2. Rooney W, Johnson G, Li X, Cohen E, Kim S, Ugurbil K, Springer C. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magnetic Resonance in Medicine 2007;57(2):308-318.
Figures