3008
Genetic impacts on nigral iron deposition in Parkinson’s disease1Department of Radiology, The Second Affiliated Hospital, Zhejiang University School of Medicine, HangZhou, China, 2Department of Radiology, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China, 3The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
Synopsis
The dysfunction of iron metabolism, especially in substantia nigra (SN), in Parkinson’s disease (PD) has been widely acknowledged, but the genetic influence on iron deposition remains largely unknown. Thus, this study aimed to explore the potential genetic impacts on iron deposition in PD patients. Using imaging genetics association analysis, this study discovers two variants, rs602201 and rs198440, have a positive impact on nigral iron deposition in PD. Specifically, patients with rs602201 polymorphism are particularly vulnerable to iron deposition in SN.
Abstract
Background: Parkinson’s disease (PD) is widely acknowledged as a complicated environmental- and genetic- related disease.(1, 2) Although the etiology of PD remains unclear, it has been well recognized that the genetic factors play an important role in disease developing processes. The mutations of many pathogenic genes have been related with both familial and sporadic PD.(2) Pathologically, excessive iron deposition in PD brain and its related oxidative damage and Lewy Body aggregation, which is closely associated with irreversible neurodegeneration, have been demonstrated as one of the main pathogeneses of PD.(3) Therefore, since the underlying mechanism for such abnormal iron metabolism remains largely unknown, the exploration, in combination of genetic examination and brain iron measurement, would be helpful to better understand the iron-related pathogenesis in PD.Iron measurement by employing MRI makes it possible to in vivo study the iron-related pathogenesis in PD. In accordance with the histopathological findings,(4) quantitative susceptibility mapping (QSM) demonstrated the excessive iron deposition in subcortical nuclei, especially in substantia nigra (SN), in PD, which indicated that QSM is a useful method to capture the iron metabolism in-vivo.
Imaging genetics is a transdisciplinary field evaluating the associations between genetic variation such as single nucleotide polymorphisms (SNPs), and imaging phenotype (also named as quantitative trait (QT)). The association studies combining neurobiological imaging are closer to the underlying neurobiology of the disease, which makes these association studies easier to identify the underlying genes.(5) However, previous researchers only explored the genetic impact of specific gene on brain alterations in PD,(6-8) which were strongly dependent on prior knowledge. Therefore, imaging genetics association framework that based on the unscreened SNP data and iron-related quantitative neuroimaging data, provides a novel approach to examine the relevant SNPs and explore the genetic impacts on the brain iron, which subsequently facilitate the clarification of iron-related pathogenesis in PD.
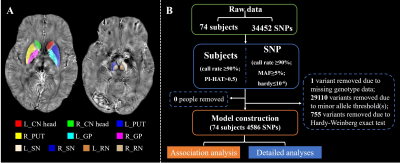
Methods: Seventy-four subjects including 38 PD patients and 36 age-matched normal controls (NC) participated in this study. ESWAN scanning, DNA detection, and basic demographic information were obtained from all subjects in “OFF” status. The tissue susceptibility of native subcortical nuclei in the basal ganglia and midbrain was extracted, including bilateral caudate head, putamen, globus palliduse, SN and red nucleus (Figure 1A). Then, the imaging genetics association analysis was used to discover the specific influence of single nucleotide polymorphism (SNP) on iron-related quantitative trait (QT) (Figure 1B). Further detailed analyses were used to illustrate the genetic effects on iron deposition at the disease level, SNP level, and their interactions.
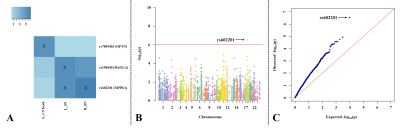
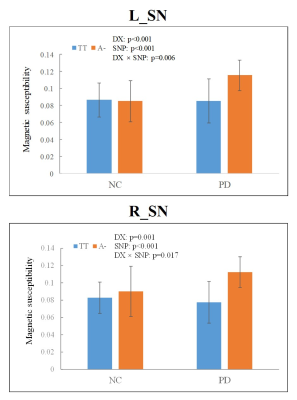
Results: Four strong SNP-QT associations were found, including rs602201 (MPPE1)-susceptibility of bilateral SNs, rs198440 (DAGLA)-susceptibility of left SN, and rs7895403 (OPTN)-susceptibility of left caudate head (Figure 2). Detailed analyses showed that (Figure 3): (1) significant iron deposition was exclusively found in bilateral SNs in PD; (2) the altered polymorphism A allele/A- genotype of rs602201 and G allele/G- genotype of rs198440 and rs7895403 were much more frequently observed in PD; (3) regarding all subjects, for rs602201, A- genotype carriers had significantly increased iron content than TT genotype in bilateral SNs; for rs198440 and rs7895403, G- carriers showed increased iron content than AA genotype in left SN and left caudate head, respectively; (4) rs602201 exhibited significant SNP-by-disease interaction in bilateral SNs.
Discussions: MPPE1 protein is a metal-dependent phosphoesterase containing mental binding and active sites, including the iron binding sites, and is widely expressed in brain, especially in the SN,(9-11) which suggests that the MPPE1 gene exerts a pivotal influence on the nigral iron deposition in PD. As for DAGLA gene, its encoded protein is a diacylglycerol lipase which is a synthesis enzyme of the endocannabinoid system (ECS).(12) The cannabinoid 1 receptor is one of the major receptors in ECS and is highly expressed in SN,(11) the signaling pathway of which is interacted with the dopaminergic D1/D2-like receptor’s,(13) suggesting that the ECS is related to the dopaminergic system and might play an important role in the pathogenesis of PD. Moreover, only PD patients showed that the altered polymorphism carriers had increased iron content compared with wild genotype subjects for both rs602201 and rs198440, which indicated that these two SNPs had specific positive effects on the nigral iron deposition in PD. In the subsequent analysis, however, a SNP-by-disease interaction effect was only observed for rs602201, which suggested that PD patients with rs602201 A- genotype may be particularly vulnerable to nigral iron deposition.
OPTN protein is an autophagy receptor and is involved in the mitophagy function.(14) Previous studies reported that OPTN variation would influence the iron metabolism by inhibiting the uptake of transferrin, resulting in the reduced cellular transferrin receptor level in cell.(15) Therefore, an interaction between OPTN protein and iron deposition was demonstrated. However, no intergroup iron deposition difference in left CN was observed, which was consistent with the former studies showing unchanged iron content in left CN between PD and NC.(16, 17) This may be on account of the region-specific iron deposition pattern in PD.
In conclusion, this study discovers two variants, rs602201 and rs198440, have a positive impact on nigral iron deposition in PD. Specifically, patients with rs602201 polymorphism are particularly vulnerable to iron deposition in SN.
Acknowledgements
This work was supported by the 13th Five-year Plan for National Key Research and Development Program of China (Grant 2016YFC1306600), the National Natural Science Foundation of China (Grant 81971577, 81701647 and 81771820), the Key Research and Development Program of Zhejiang Province (Grant 2020C03020), the Zhejiang Provincial Natural Science Foundation (Grant LQ20H180012), the China Postdoctoral Science Foundation (Grant 2019M662082). We thank all patients with Parkinson’s disease and normal controls who participated in this study.References
1. Lees AJ, Hardy J, Revesz T (2009): Parkinson's disease. Lancet. 373:2055-2066.
2. Gasser T (2003): Overview of the genetics of parkinsonism. Adv Neurol. 91:143-152.
3. Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L (2014): The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 13:1045-1060.
4. Langkammer C, Schweser F, Krebs N, Deistung A, Goessler W, Scheurer E, et al. (2012): Quantitative susceptibility mapping (QSM) as a means to measure brain iron? A post mortem validation study. Neuroimage. 62:1593-1599.
5. Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. (2009): Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS One. 4:e6501.
6. Thaler A, Artzi M, Mirelman A, Jacob Y, Helmich RC, van Nuenen BF, et al. (2014): A voxel-based morphometry and diffusion tensor imaging analysis of asymptomatic Parkinson's disease-related G2019S LRRK2 mutation carriers. Mov Disord. 29:823-827.
7. Zhang K, Tang Y, Meng L, Zhu L, Zhou X, Zhao Y, et al. (2019): The Effects of SNCA rs894278 on Resting-State Brain Activity in Parkinson's Disease. Front Neurosci. 13:47.
8. Nombela C, Rowe JB, Winder-Rhodes SE, Hampshire A, Owen AM, Breen DP, et al. (2014): Genetic impact on cognition and brain function in newly diagnosed Parkinson's disease: ICICLE-PD study. Brain. 137:2743-2758.
9. Lohoff FW, Ferraro TN, Brodkin ES, Weller AE, Bloch PJ (2010): Association between polymorphisms in the metallophosphoesterase (MPPE1) gene and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 153B:830-836.
10. Reiter TA, Rusnak F (2004): Electrochemical studies of the mono-Fe, Fe-Zn, and Fe-Fe metalloisoforms of bacteriophage lambda protein phosphatase. Biochemistry. 43:782-790.
11. Vuoristo JT, Ala-Kokko L (2001): cDNA cloning, genomic organization and expression of the novel human metallophosphoesterase gene MPPE1 on chromosome 18p11.2. Cytogenet Cell Genet. 95:60-63.
12. Sanchez-Marin L, Gavito AL, Decara J, Pastor A, Castilla-Ortega E, Suarez J, et al. (2020): Impact of intermittent voluntary ethanol consumption during adolescence on the expression of endocannabinoid system and neuroinflammatory mediators. Eur Neuropsychopharmacol. 33:126-138.
13. Ranieri R, Laezza C, Bifulco M, Marasco D, Malfitano AM (2016): Endocannabinoid System in Neurological Disorders. Recent Pat CNS Drug Discov. 10:90-112.
14. Weil R, Laplantine E, Curic S, Genin P (2018): Role of Optineurin in the Mitochondrial Dysfunction: Potential Implications in Neurodegenerative Diseases and Cancer. Front Immunol. 9:1243.
15. Sirohi K, Chalasani ML, Sudhakar C, Kumari A, Radha V, Swarup G (2013): M98K-OPTN induces transferrin receptor degradation and RAB12-mediated autophagic death in retinal ganglion cells. Autophagy. 9:510-527.
16. Guan X, Xuan M, Gu Q, Huang P, Liu C, Wang N, et al. (2017): Regionally progressive accumulation of iron in Parkinson's disease as measured by quantitative susceptibility mapping. NMR Biomed. 30.
17. He N, Ling H, Ding B, Huang J, Zhang Y, Zhang Z, et al. (2015): Region-specific disturbed iron distribution in early idiopathic Parkinson's disease measured by quantitative susceptibility mapping. Hum Brain Mapp. 36:4407-4420.
Figures