2997
Fully Automated Pelvic Bones Segmentation in Multiparameter MRI Using a 3D Convolutional Neural Network1department of radiology, peking university first hospital, Beijing, China
Synopsis
This retrospective study aims to perform automated pelvic bones segmentation in multiparametric MRI (mpMRI) using 3D convolutional neural network (CNN). 264 pelvic DWI images and corresponding ADC maps obtained from three MRI vendors from 2018 to 2019 were used for the 3D U-Net CNN development. 60 independent mpMRI data from 2020 were used to externally evaluate the segmentation model using quantitative criteria (Dice similarity coefficient) and qualitative assessment (SCORE system). The results demonstrated that the 3D CNN can achieve fully automated pelvic bone segmentation on multi-vendor DWI and ADC images with good quantitative and qualitative performances.
Introduction
Accurate segmentation of pelvic bones is considered as the first step to achieve accurate detection and localization of pelvic bone metastases1. However, the ill-defined boundary among bone structures posed a challenge to the segmentation.Objective
To present a deep learning-based approach for automated pelvic bones segmentation in multiparametric MRI (mpMRI) using 3D convolutional neural network (CNN).Methods
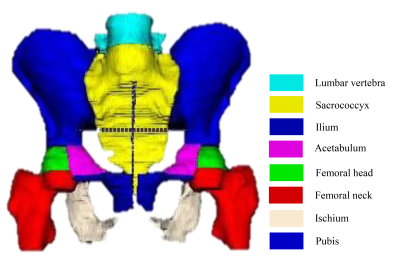
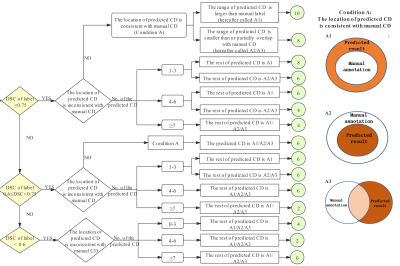
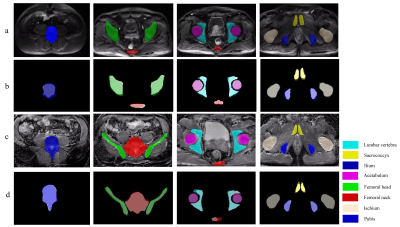
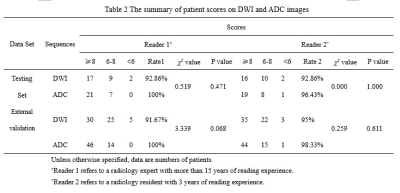
This IRB-approved retrospective study included 264 pelvic mpMRI data obtained from three MRI vendors from 2018 to 2019. The manual annotations of pelvic bones (included lumbar vertebra; sacrococcyx; ilium; acetabulum; femoral head; femoral neck; ischium; pubis) on DWI and ADC images were used to create reference standards (Fig. 1). A 3D U-Net CNN was employed for automatic pelvic bones segmentation2. 60 independent mpMRI data from 2020 were additionally used to externally evaluate the model. The segmentation results were assessed quantitatively to the reference standards using the Dice similarity coefficient (DSC). To evaluate whether the predicted results meet the requirements of clinical application. A SCORE system established by us (Fig. 2) was used to qualitatively evaluate the model by two readers, and a rate of up-to-standard >80% was considered to meet the demand of clinical application.Results
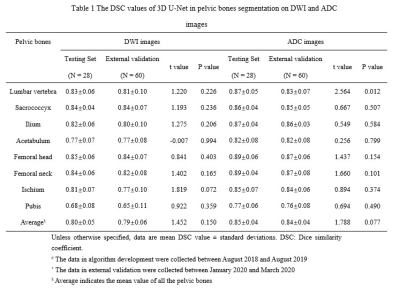
The 3D U-Net achieved high average DSC in both testing and external validation set (testing set: 0.80 [DWI images] and 0.85[ADC images]; external validation set: 0.79 [DWI images] and 0.84 [ADC images]) as shown in Table 1 and Figure 3. In terms of qualitative evaluation, all of the rates of up-to-standard in both testing and external validation set are highly above 80%, and intraclass correlation coefficient (ICC) of the SCORE results between two readers was 0.842 (95% confidence interval: 0.793-0.880) (Table 2).Discussion
In this research, the quantitative and qualitative evaluations were used to determine the value of clinical application of the 3D U-Net model. The CNN achieved good quantitative segmentation performance on pelvic bones with high average DSC values on DWI images and ADC images in both testing set and external validation set. More importantly, all of the rates of up-to-standard in both DWI and ADC images are highly above 80%, which demonstrated the good acceptability of the 3D U-Net in clinical practice. Besides, the excellent concordance between two readers (ICC = 0.842) further confirmed the feasibility of clinical application of this CNN. Our research may provide essential geometric information for subsequent detection of pelvic bone metastases.Conclusion
A deep learning-based method can achieve fully automated pelvic bone segmentation on multi-vendor DWI and ADC images with good quantitative and qualitative performances.Acknowledgements
The authors gratefully acknowledge the technical support of Yaofeng Zhang, Dadou Zhang and Jiahao Huang from Beijing Smart Tree Medical Technology Co. Ltd.References
1. Lindgren Belal S, Sadik M, Kaboteh R, et al. Deep learning for segmentation of 49 selected bones in CT scans: First step in automated PET/CT-based 3D quantification of skeletal metastases. Eur J Radiol 2019;113:89-95.
2. Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O: 3D U-net: Learning dense volumetric segmentation from sparse annotation. In: Int conf Image Comput Comput Interv. 2016: 424–432.
Figures