2992
Model-based Deep Learning Reconstruction using Folded Image Training Strategy (FITS-MoDL) for Liver MRI Reconstruction
Satoshi Funayama1,2, Utaroh Motosugi3, and Hiroshi Onishi1
1Department of Radiology, University of Yamanashi, Yamanashi, Japan, 2Graduate School of Medicine, University of Yamanashi, Yamanashi, Japan, 3Department of Radiology, Kofu-Kyoritsu Hospital, Yamanashi, Japan
1Department of Radiology, University of Yamanashi, Yamanashi, Japan, 2Graduate School of Medicine, University of Yamanashi, Yamanashi, Japan, 3Department of Radiology, Kofu-Kyoritsu Hospital, Yamanashi, Japan
Synopsis
Short acquisition time is one of the key features of liver MRI to acquire images during breath-holding. A combination of undersampling and deep learning-based reconstruction would be a powerful reconstruction method to achieve sufficient speed and SNR. However, it is challenging due to high memory consumption in network training. The folded image training strategy (FITS) is one of the methods to handle this problem. In this study, we demonstrated that the model-based deep learning reconstruction using FITS had better image quality in liver MRI acquired with multiple coils.
Introduction
Model-based deep learning reconstruction which combines image prior estimated by convolutional neural network (CNN) and conventional model-based reconstruction (e.g., parallel imaging) showed better image quality.1–3 These networks, however, requires high memory consumption during training, which leads many limitations; input image size (acquisition matrix), depth of network, and batch size in network training. To handle this problem, we previously reported a network training with FITS.4 FITS reduced memory consumption during network training effectively in single coil model. However, feasibility for multi coil model-based case has been still unclear. The purpose of this study was to demonstrate a feasibility of FITS in multi coil model-based deep learning reconstruction in liver MRI.Methods
Image Reconstruction: The k-space data were retrospectively undersampled to compare the utility of 3 different reconstruction methods: FITS-MoDL, conventional MoDL (conv-MoDL), and total variation regularized compressed sensing (CS). The undersampling masks were created using Gaussian random sampling. The center of the masks was fully sampled as auto-calibration area (24 x 16) for ESPIRiT sensitivity estimation.5 For computational benefit, principal component analysis-based coil compression was applied.6Network Architecture: The proposed FITS-MoDL is an unrolled model-based deep learning reconstruction which iterates 2D CNN block and data consistency block to utilize the fidelity of sampled k-space point through the iterations (Figure 1). This architecture has two times deeper 2D CNN block (10 layers) compared to conventional MoDL. To train this deeper network, images for network training were folded along phase encoding direction using uniform undersampling by two, which made memory consumption small (Figure 2). After the network training, images which had full size along phase encoding direction were used for the network inference. For comparison, conventional MoDL network which had 5 layers in the 2D CNN block was trained with same dataset. All images were reconstructed in y-z plane. These networks were trained end-to-end based on the mean squared error between output and ground truth images using Adam optimizer (learning rage: 1.0x10-3) on a NVIDIA TITAN RTX graphics accelerator.
Training Data: Clinical upper abdominal T1-weighted images of 122 patients were used for this study. The images were fully sampled using a 3.0-T scanner (Discovery 750, GE Healthcare) and 32-channel torso coil. The acquisition parameters were as follows: TR = 3.9 ms, TE = 2.2 ms, FA = 15°, field-of-view = 360 × 360 mm2, receiver bandwidth = ±166.67 kHz, slice thickness/intersection gap = 3.6/0 mm, matrix = 288 × 192, parallel imaging factor (phase x slice) = 1.5 × 1, acquisition time = 20 s, using an elliptical scan (Turbo mode). These images were randomly divided into training set (n=40), validation set (n=5), and test set (n=77). Data augmentation was applied using multiple acceleration factor (4 and 6) of undersampling mask. Finally, 18880 images were used for the network training.
Evaluation: Image quality of reconstructed images were evaluated using test set in terms of peak signal-to-noise ratio (PSNR) and structure similarity index (SSIM). Acceleration factors of 2, 4, and 6 were used to build undersampling masks for evaluation. Paired t-test was used to compare the metrics differences between FITS-MoDL and the others. Bonferroni correction was used to handle multiple comparison.
Results
The proposed network trained with FITS showed the best quality in all acceleration factors compared with conv-MoDL and CS (Figure 3). In acceleration factor of 4, FITS-MoDL had significantly higher PSNR and SSIM compared with the others: PSNR, FITS-MoDL 46.10±1.67 (dB, mean±SD, reference) vs. conv-MoDL 45.94±1.78 (p<0.001), CS 43.84±1.87 (p<0.001). The memory consumption during network training for FITS-MoDL (1535 MiB) was less than that for conv-MoDL (1795 MiB). A representative case was shown in figure 4.DIscussion
The FITS-MoDL outperformed the conventional network in terms of image quality and memory consumption during network training. This result is consistent with the previous report done with single coil settings, i.e. FITS works well not only for single coil case but also for multi coil case. In this study, we used the extra-memory capacity gained by FITS to make the network deeper. The extra-memory capacity by FITS can be used for other purposes (e.g., larger acquisition matrix), because FITS does not restrict network architecture or acquisition parameters.A limitation of this study could be the lack of visual assessment of images by radiologists. Further study is required to investigate diagnostic benefit and a risk of artifacts of deep learning reconstruction which some authors mentioned.
Acknowledgements
No acknowledgement found.References
- Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79(6):3055-3071.
- Schlemper J, Caballero J, Hajnal J V., Price AN, Rueckert D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging. 2018;37(2):491-503.
- Aggarwal HK, Mani MP, Jacob M. MoDL: Model-Based Deep Learning Architecture for Inverse Problems. IEEE Trans Med Imaging. 2019;38(2):394-405.
- Funayama S, Wakayama T, Onishi H, Motosugi U. FITs-CNN: A Very Deep Cascaded Convolutional Neural Networks Using Folded Image Training Strategy for Abdominal MRI Reconstruction. In: Proc. Intl. Soc. Mag. Reson. Med. ; 2020:1004.5.
- Uecker M, Lai P, Murphy MJ, et al. ESPIRiT - An eigenvalue approach to autocalibrating parallel MRI: Where SENSE meets GRAPPA. Magn Reson Med. 2014;71(3):990-1001.
- Huang F, Vijayakumar S, Li Y, Hertel S, Duensing GR. A software channel compression technique for faster reconstruction with many channels. Magn Reson Imaging. 2008;26(1):133-141.
Figures
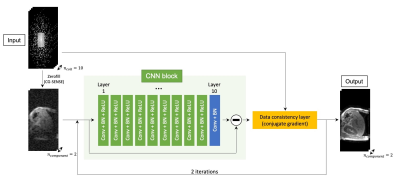
Overview of proposed network architecture (FITS-MoDL)
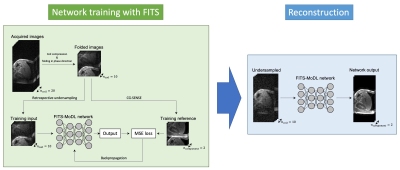
Flow diagram of network training with folded image training strategy (FITS) and image reconstruction. In training with FITS, images for training were folded by factor of 2 to reduce memory consumption.

PSNR and SSIM of model-based deep learning reconstruction (FITS-MoDL), conventional model-based deep learning reconstruction (conv-MoDL), and total variation regularized compressed sensing (CS). The metrics were evaluated for acceleration factor of 2, 4, and 6. The FITS-MoDL showed significantly higher image quality compared to the other two methods (p<0.001).
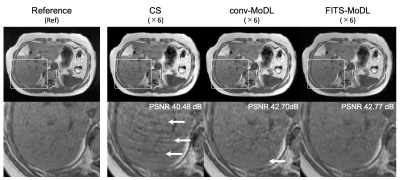
A representative case. (CS) total variation regularized compressed sensing showed some aliasing on liver parenchyma. A few aliasing is remained in the conventional model-based deep learning reconstruction (conv-MoDL), while it is removed in the model-based deep learning reconstruction using FITS (FITS-MoDL).