2990
Deep Learning 3D Convolutional Neural Network for Noninvasive Evaluation of Pathologic Grade of HCC Using Contrast-enhanced MRI1The First Affiliated Hospital of Dalian Medical University, Dalian, China, 2Chengdu Institute of CoChinese Academy of Sciences, Chengdu, China, 3University of Chinese Academy of Sciences, Beijing, China, 4GE Healthcare (China), Shanghai, China
Synopsis
In recent years, convolutional neural networks (CNNs) have become one of the most advanced deep learning networks. Deep learning with CNNs has reportedly achieved good performance in the pattern recognition of images. In the present study, 3D-CNN based on contrast-enhanced (CE)-MR images was demonstrated to be capable to evaluate pathologic grade of hepatocellular carcinoma (HCC) treated with surgical resection, which will provide more prognostic information and facilitate clinical management.
Purpose
To investigate the diagnostic performance of our proposed three-dimensional convolutional neural network (3D-CNN) model for the differentiation of the pathologic grade of hepatocellular carcinoma (HCC) based on contrast-enhanced (CE)-MR images.Introduction
HCC, the most common primary malignant liver tumor, is the second most common cause of death related to malignancy in the world [1]. Although the surgical resection of HCC has been improved, patient prognosis remains poor due to the high recurrence rate. The pathologic grade of HCC is one of the most important factors in evaluating early recurrence after surgical resection [2]. Compared to well-differentiated (WD) or moderately differentiated (MD) HCCs, poorly differentiated (PD) HCC has a poorer prognosis and higher tumor recurrence. PD HCC is also associated with a worse survival rate than WD or MD HCCs [3]. The development of noninvasive imaging techniques to safely and accurately assess the pathologic grade of HCC would benefit the selection of an optimal treatment method for patients and improve their survival rate. In recent years, CNNs have become one of the most advanced deep learning networks. Deep learning with CNNs has reportedly achieved good performance in the pattern recognition of images [4]. Some preliminary achievements of computer-aided diagnostic techniques based on CNNs have also been obtained in medical image analysis for the detection, segmentation, and grading of abdominal lesions in a broad spectrum of diseases [5-8]. In this study, we used a 3D-CNN to extract temporal sequence information and spatial texture information from CE-MR images.Methods
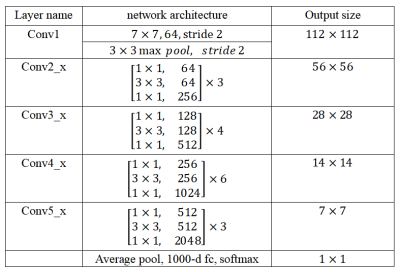
This retrospective study enrolled 113 patients who were pathologically confirmed as HCC, including 60 PD HCCs and 53 non-PD (WD and MD) HCCs. All patients have underwent preoperative CE-MR examinations within two weeks before resection. The samples were randomly divided into the training set (n = 90) and the testing set (n = 23) at ratio of 8:2. Figure 1 shows the flowchart of the multimodal fusion model. On the arterial (A), venous (V) and delayed (D) phase images, two radiologists manually outlined the regions of interest (ROIs) which enclosed the boundary of the lesions. In this paper, we set the size of the normalized HCC slice as 224 × 224 to input into the network. To reveal the characteristics of the temporal and spatial information of the CE-MR images to aid HCC diagnosis, the ResNet-50 was used to extract senior semantic features based on each phase images. The ResNet-50 consisted of a 64-dimensional convolutional layer (7×7), a Max pooling layer (3×3), four bottleneck architectures, an average pooling layer, and a fully connected layer with 1000 neural elements (shown in Table 1). Figure 2 shows the feature extraction process of ResNet-50. In order to adapt the feature representation ability of CNN in public image dataset to our model, we used the parameters of ResNet-50 model trained on ImageNet dataset to initialize our three ResNet-50 model. In order to make the ResNet-50 network on different phase images to better learn to express the characteristics of HCC differentiation, we deleted the last full connected layer of each ResNet-50, and then added two fully connected layers with 2048 and two neurons respectively. Finally, we added two fully connected layers followed by the three models (A, V and D). These two fully connected layers were used to fuse the senior semantic features of different phase MR images for the HCC classification task. The area under the curve (AUC), accuracy, sensitivity, and specificity were used to evaluate the diagnostic performance for differentiation of pathologic grade of HCC.Results
Table 2 shows the comparison of performances between the single-modal model and multimodal fusion model in the testing set. The multimodal fusion model showed prior performance (AUC = 0.889, sensitivity = 100.0%, specificity = 77.8%) compared with the single-modal model (A model: AUC = 0.579, sensitivity = 78.1%, specificity = 37.7%; V model: AUC: 0.793, sensitivity = 80.0%, specificity = 78.6%; D model: AUC = 0.681, sensitivity = 46.4%, specificity = 89.8% ).Discussion and Conclusion
Poor differentiation of HCC has been proved as a risk factor of bad prognosis, and it can only be accurately identified by pathology. This pilot study indicated that the 3D-CNN model may be valuable for the noninvasive evaluation of the pathologic grade of HCC; however, further study would be necessary to invalidate our model.Acknowledgements
NoneReferences
[1] Tang Y, Wang H, Ma L, et al. Diffusion-weighted imaging of hepatocellular carcinomas: a retrospective analysis of correlation between apparent diffusion coefficients and histological grade. Abdom Radiol (NY). 2016;41(8):1539-45.
[2] Kim DJ, Clark PJ, Heimbach J, et al. Recurrence of hepatocellular carcinoma: importance of mRECIST response to chemoembolization and tumor size. Am J Transplant. 2014;14(6):1383-90.
[3] Yang DW, Jia XB, Xiao YJ, et al. Noninvasive Evaluation of the Pathologic Grade of Hepatocellular Carcinoma Using MCF-3DCNN: A Pilot Study. Biomed Res Int. 2019;2019:9783106.
[4] Shin HC, Roth HR, Gao M, et al. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging. 2016; 35(5): 1285-1298.
[5] Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60-88.
[6] Qi Dou, Hao Chen, Lequan Yu, et al. Automatic Detection of Cerebral Microbleeds From MR Images via 3D Convolutional Neural Networks. IEEE Trans Med Imaging. 2016;35(5):1182-1195.
[7] Lemaître G, Martí R, Freixenet J, et al. Computer-Aided Detection and diagnosis for prostate cancer based on mono and multi-parametric MRI: a review. Comput Biol Med. 2015;60:8-31.
[8] Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61-78.