2989
A Two-Stage Deep Learning Model for Accurate Vessel Segmentation and Reconstruction in the MRI of Live1Chengdu Institute of Computer Application, Chinese Academy of Sciences, Chengdu, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Department of Radiology, the First Affiliated Hospital of Dalian Medical University, Dalian, China
Synopsis
The deep learning technology was used on the segmentation automatically and reconstruction of the intrahepatic portal vein to get its volume and three-dimensional spatial position information. The results showed that the dice accuracy was 83.32% in the training and 75.89% in the test set among 12 cases. The current study demonstrated the volume of intrahepatic vessels and 3D spatial location information can be obtained by the tow-stage deep learning model and it can be applied in promising clinical effectively.
Purpose
Volume and spatial location of intrahepatic vessels are very important information for interventional surgery of liver tumors. However, it is difficult to give direct and accurate answers to these questions depending on the information of human liver given by traditional MRI images.Introduction
The liver is a metabolic organ of human body and have a complex internal plumbing system. The dual blood supply of liver involved portal vein (about 70%) and hepatic artery (only 30%). Arterial blood enriched oxygen and nutrients flowing in hepatic artery same as the artery in other organs. The blood flowing in portal vein mainly comes from venous blood system of digestive tract. The branches of hepatic artery and portal vein around with few small connective tissue become more and more thin and parallel with the bile duct after entering the liver from the hepatic portal. Its every significant to the follow-up clinical decision-making to determine the volume of intrahepatic blood vessels and the location of blood vessels and tumors. In recent years, deep convolution neural network has got great progress in the organ segmentation and curative effect prediction. This is a worthy of we think deeply and investigate of problem that can we obtain the volume information and 3D spatial position information of vessels by reconstructing image sequences followed extract blood vessels from 2D MRI images using deep convolution neural network.Materials and Methods
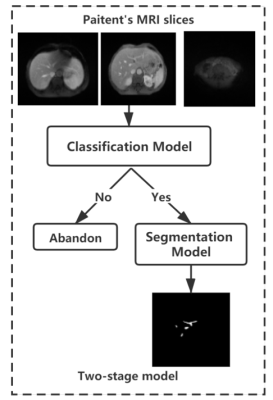
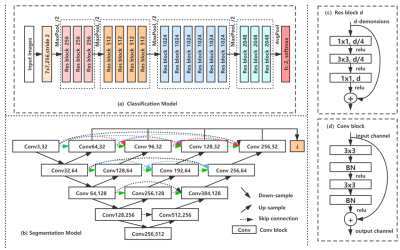
In this this study we collected respectively 12 patients with portal phase MRI images from the first people's Hospital Affiliated to Dalian Medical University.The MRI images were reviewed by two radiologists and the portal vein vessels on each slice of the patient were manually sketched on itk-snap. the final consensus verdict will be given after discussion if two radiologists have disparity with each other. We used a two-stage approach since the complex distribution of small blood vessels in the liver different from other organ segmentation(Figures 1 and 2). Firstly, a classification network ResNet was used to check for the presence of portal vein vessels in the current slices1. ResNet, a simple but efficient classification network,can increase the flow of information between different layers by residual blocks. Then an UNet++ network was used to segment the selected slices. UNet++ is tried and true network method in many area especially the field of medical image2. And the UNet++ adopts the structure of encoding and decoding and skip connection to save the details. At the same time, we also compared the results of segmentation direct using UNet++ network and 3D Vnet3. Finally, VTK was used to reconstruct the 2D segmentation results of intrahepatic portal vein4. The volume of intrahepatic portal vein is acquired though combine the voxel information and the total number of vascular pixels of each slice calculated by the segmentation results of intrahepatic portal vein obtained in the previous stage. Simultaneously, the 3D structure of intrahepatic portal vein can be reconstructed according to the segmentation results of intrahepatic portal vein and voxel information.Results
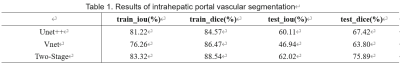
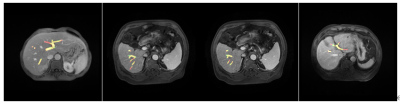
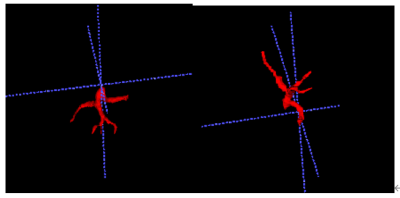
The result showed that the performance of the proposed two-stage split network is the best and its IOU and dice were higher than UNet++ and VNet(Table.1). The visualization results demonstrated segmentation results of network are very close to the result by doctors outlining(Fig.3). In the work of vascular volume measurement, we compared the results with the radiologist because there is no way to obtain the real intrahepatic portal vein volume of the patient. And the result claimed our method also superior to other methods. We reconstructed the 3D results(Fig. 4).Discussion
The two-stage segmentation network can effectively reduce light over segmentation phenomenon.But we only delineated the intrahepatic portal vein in this experiment and the depiction of other intrahepatic blood vessels should be involved in further study.Conclusion
Its crutial to get reliable basis for subsequent clinical treatment especially the three-dimensional spatial information and volume information of intrahepatic portal vessels. For that reason, we suggested the two-stage vascular segmentation method are more expressive than trandional MRI images.Acknowledgements
Supported by grants from the National Natural Science Foundation of China (6197010131).References
1. He, Kaiming , et al. "Deep Residual Learning for Image Recognition." IEEE Conference on Computer Vision & Pattern Recognition IEEE Computer Society, 2016.
2. Zhou, Zongwei, et al. "Unet++: A nested u-net architecture for medical image segmentation." Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Springer, Cham, 2018. 3-11.
3. Milletari, Fausto, Nassir Navab, and Seyed-Ahmad Ahmadi. "V-net: Fully convolutional neural networks for volumetric medical image segmentation." 2016 fourth international conference on 3D vision (3DV). IEEE, 2016.
4. Schroeder, William J., Lisa Sobierajski Avila, and William Hoffman. "Visualizing with VTK: a tutorial." IEEE Computer graphics and applications 20.5 (2000): 20-27.
Figures