2988
Deep learning-based detection of liver disease using MRI
Mark A Pinnock1,2, Yipeng Hu1,2, Alan Bainbridge3, David Atkinson4, Rajeshwar P Mookerjee5, Stuart A Taylor4, Dean C Barratt1,2, and Manil D Chouhan4
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Wellcome/EPSRC Centre for Interventional and Surgical Sciences, University College London, London, United Kingdom, 3Department of Medical Physics and Biomedical Engineering, University College London Hospitals NHS Foundation Trust, London, United Kingdom, 4Centre for Medical Imaging, Division of Medicine, University College London, London, United Kingdom, 5Institute for Liver and Digestive Health, Division of Medicine, University College London, London, United Kingdom
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Wellcome/EPSRC Centre for Interventional and Surgical Sciences, University College London, London, United Kingdom, 3Department of Medical Physics and Biomedical Engineering, University College London Hospitals NHS Foundation Trust, London, United Kingdom, 4Centre for Medical Imaging, Division of Medicine, University College London, London, United Kingdom, 5Institute for Liver and Digestive Health, Division of Medicine, University College London, London, United Kingdom
Synopsis
Traditional approaches to MRI detection of liver disease require specialist hardware, sequences and post-processing. Here we propose a deep learning (DL) based model for the detection of liver disease using standard T2-weighted anatomical sequences, as an early feasibility study for the potential of DL-based classification of liver disease severity. Our DL model achieved a diagnostic accuracy of 0.92 on unseen data and achieved a test accuracy of 0.75 when trained with relevant anatomical segmentation masks without images, demonstrating potential scanner/sequence independence. Lastly, we used DL interpretability techniques to analyse failure cases.
Introduction
Liver disease is a major cause of morbidity and mortality worldwide1. Quantitative MRI methods examining the liver, such as MR elastography and proton-density fat fraction sequences, can quantify fibrosis and steatosis2 but require additional hardware, sequences and post-processing.Deep learning using non-quantitative anatomical data has garnered much attention in medical imaging, with applications including detection of liver tumours, fibrosis and organ segmentation3.
As an initial step toward developing a tool to evaluate liver disease severity, we aim in this study to: 1) develop and evaluate the use of a convolutional neural network (CNN) for the classification of healthy subjects versus those with liver disease from anatomical T2-weighted MR imaging; 2) assess model accuracy and the effect of the inclusion of relevant anatomical segmentation masks; 3) evaluate model portability through the use of segmentation mask data alone and; 4) explore model failure cases using CNN interpretability techniques.
Methods
PatientsEthical approval was obtained and all participants provided prospective informed written consent. Healthy volunteers (n=13) and patients with histologically confirmed but stable cirrhosis (n=9 Child-Pugh score A; n= 9 Child-Pugh score B) were recruited.
MR Imaging
Scans were obtained using a 3.0T scanner (Achieva, Philips Healthcare, Best, Netherlands) with a 16-channel body coil. A breath-hold balanced steady-state free precession (SSFP) sequence was used for anatomical coronal imaging centred on the liver (TR=2.5ms, TE=1.2ms, α=45°, matrix size 352x352 pixels, field-of-view=350x350mm, slice thickness=5mm). Manual segmentation of the liver and spleen (3 slices per organ) and all imaged splanchnic vessels was performed by an abdominal radiologist.
Transfer learning-based CNN
The CNN used was based on MobileNetV24, available from Tensorflow5 already pretrained on ImageNet6. Top classification layers were replaced with a dense layer of size 128 and a dense layer of size 1 with sigmoid output to form a binary classification head. Prior to training, the images and masks were resized to 224x224 with normalisation of pixel intensities.
Study protocol and statistical analysis
Scans were randomised into training (n=8 healthy volunteers, n=11 patients) and testing (n=5 healthy volunteers, n=7 patients) groups. Three models were trained with (1) segmentation masks only, (2) images only and (3) images and their respective segmentation masks. Diagnostic accuracy for the presence of liver disease was assessed by comparing sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy and F1 scores. Mis-classified cases in the test cohort were examined using GradCAM7 for the model trained on images/images with segmentation masks. This technique visualises the activations contributing to the predicted class.
Results
Model developmentThe network was trained with frozen pre-trained weights for 50 epochs on a Nvidia GTX 1050Ti 4Gb graphics card using the Adam optimiser (learning rate 1e-4 , beta1 = 0.9, beta2 = 0.999) before fine-tuning the top 50 layers for 50 further epochs at learning rate 1e-5. The minibatch size was fixed at 32 and the loss function used was binary cross-entropy.
Model accuracy
Table 1 shows subject-level diagnostic accuracy metrics for the three models. The image-trained models misclassified one subject as positive, while adding the segmentation masks to the images during training demonstrated no improvement.
Model portability
The CNN trained using segmentation masks only was less accurate than the image-based models (kappa = 0.625), with low specificity (0.60) but an overall accuracy of 0.75.
Model interpretability
Figures 1 and 2 show representative GradCAM visualisations for incorrect and correct classifications from the image-only model. Table 2 shows the percentages of test images that display strong activations in the liver, spleen and mediastinum in image-level true and false positives. Correct classifications tend to be focused on the liver and spleen, however in false positives the mediastinum becomes more relevant in the false positive images while the spleen becomes less important. When segmentation masks are included in training, the mediastinum contributes substantially less to false positive classifications.
Discussion
We have employed deep learning to classify liver disease from anatomical MRI images using a small number of training examples. Adding limited segmentation masks to the images did not affect the overall classification accuracy, but did alter the anatomical regions contributing to classifications. Using the masks alone had inferior accuracy – however with a larger training dataset it may be feasible to increase this accuracy enough to perform classification independently of different institution scanners. Lastly, we have explored the use of CNN interpretability tools to show how the image-only model heavily weights the mediastinum in its false positive classifications, an effect overcome when relevant anatomical segmentation masks are included in training.This is a small scale study and while the generated models are promising, further research is planned to explore how well they generalise to larger datasets.
Acknowledgements
No acknowledgement found.References
- Asrani SK et al. Burden of liver diseases in the world. Journal of hepatology 70.1: 151-171 (2019)
- Castera L et al. Noninvasive assessment of liver disease in patients with nonalcoholic fatty liver disease. Gasteroenterology 156.5 (2019): 1264-1281
- Zhou L, et al. Artificial intelligence in medical imaging of the liver. World journal of gastroenterology 25.6 (2019): 672
- Sandler M, et al. Mobilenetv2: Inverted residuals and linear bottlenecks. Proceedings of the IEEE conference on computer vision and pattern recognition (2018)
- Abadi M et al. Tensorflow: Large-scale machine learning on heterogenous distributed systems. arXiv preprint 1603.04467 (2016)
- Deng J et al. Imagenet: A large-scale hierarchical image database. IEEE conference on computer vision and pattern recognition (2009)
- Selvaraju RR et al. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE conference on computer vision (2017)
Figures
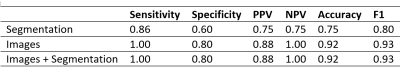
Table 1: Subject level diagnostic metrics for models trained on segmentations only, images only, and images with segmentations
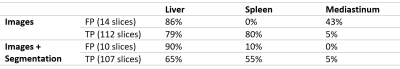
Table 2: Percentage of false positive (FP) and true positive (TP) images containing strong activations in liver, spleen and mediastinum
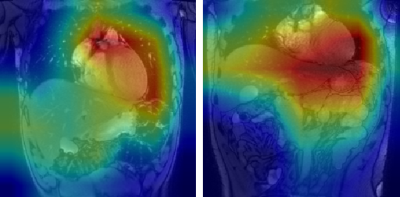
Figure 1: GradCAM visualisations for two false positive classifications
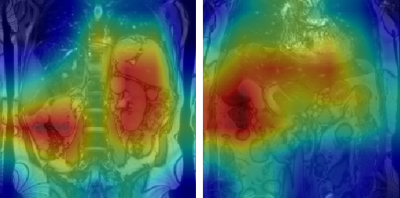
Figure 2: GradCAM visualisations for two true positive classifications