2969
Wrist cartilage segmentation using U-Net convolutional neural networks1Department of Physics and Engineering, ITMO University, Saint Petersburg, Russian Federation
Synopsis
Segmentation of wrist cartilage may be of interest for the detection of cartilage loss during osteo- and rheumatoid arthritis. In this work, U-Net convolutional neural networks were used for automatic wrist cartilage segmentation. The networks were trained on a limited amount of labeled data (10 3D VIBE images). The results were compared with the previously published for a planar patch-based archutecture (3D DSC = 0.71). Utilisation of U-Net archutecture and data augmentation allowed to significantly increase the segmentation accuracy in lateral slices. Truncation of the deepest level in the classical U-Net architecture provided the 3D DSC=0.77.
Introduction
Segmentation of cartilage in MR images has been reported to be useful for the detection of cartilage loss during rheumatoid arthritis and osteoarthritis. Convolutional neural networks (CNNs) have been recently proposed as an efficient and fully automatic tool for this purpose[1]. Plenty of architectures and approaches have been utilized for cartilage segmentation in knee joint images. For a more subtle and complex wrist joint only one planar CNN architecture[2], which provided a sufficient accuracy only for medial cross-sections of the wrist joint, was proposed. This planar CNN outperformed a state-of-the-art U-Net architecture[3] when trained on a limited amount of data. In this study, we investigate if the optimization of U-Net architecture and learning parameters and augmentation of data may gain the wrist cartilage segmentation accuracy and allow automatic measurement of wrist cartilage.Methods
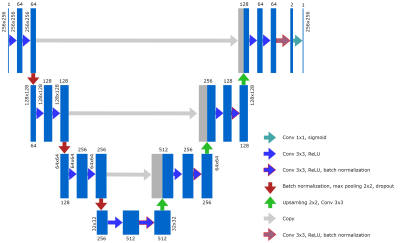
In this research, two CNN architectures were used for automatic wrist cartilage segmentation. The first CNN was based on a classical image-based U-Net[3]. Batch normalization layers were added to the architecture in order to reduce training time; dropout and noise layers were added to avoid overfitting. Such parameters as the probability of dropouts, noise level, learning rate, and batch size were optimized in a grid search. As a second option, we considered the U-Net-based network with a truncated layer that made the CNN less deep but at the same time reduced the number of trainable parameters (8.6*106 vs 34.5*106 for the classical U-Net) that is usually beneficial for training on a limited amount of training data. The truncated U-net was trained with the same parameters as the classical one except for the probability of dropouts and noise level, which were adjusted.The dataset containing 10 partly labeled (every second 2D slice was labeled) 3D VIBE wrist images (FOV = 97 x 120 mm2, matrix size = 260 x 320, 284 planar coronal images in total) that were previously used for training a planar patch-based (PB) CNN[2] was used for CNNs training in this research. The planar images were cropped to size 256x256 and augmented with library albumentations. Four types of transformations were used: vertical flip, horizontal flip, arbitrary angle rotation, elastic transform, and grid distortion. Each of the transformations had its own probability of occurrence. The augmentation provided a training dataset of 1136 images. 7% of these images were used for the validation of the training process. Another set of 10 3D images (contained 260 planar images) was used for testing.
CNNs were built using Python 3.6.4, TensorFlow 1.7.0, and Keras 2.1.5 open-source neural network library. The training was performed on a Tesla workstation with Nvidia Tesla P100 GPU, the testing was performed on a PC with Nvidia GTX 1050 GPU.
Cartilage masks provided by the CNNs for the test dataset were compared with the corresponding manual labels. Corresponding Dice similarity coefficients (DSC) were calculated for each of the 3D images as a whole and on the layer-to-layer basis as was previously proposed[2]: an average DSC was calculated for each of 4 zones: zone #1 encompassed slices with no cartilage, zone #2 encompassed lateral slices with a relative amount of cartilage (relatively to a slice of this 3D image with the maximal amount of cartilage) from 0 to 33%, zone#3 - from 34% to 66%, and zone#4 - from 67% to 100%. These values were compared with the corresponding results for a reference PB-CNN[3]. The performance for medial slices was compared to the reported results of the manual segmentation procedure[2].
Results and Discussion
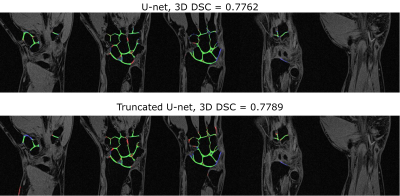
The optimal learning parameters, which provided the highest validation DSC during training for the classical U-Net, were the following: batch size - 32, learning rate – 0.5e-3, noise level - 0.3, probability of dropouts – 0.4. For the truncated U-Net, the optimal noise level was 0.2 and the probability of dropouts was 0.35. The architecture of the truncated U-Net is depicted in Fig.1.The results of the layer-to-layer performance of the used U-Net-based networks and of the reference one are summarized in the table in Fig.2. Interestingly, the performance of both U-Nets was higher than of a PB-CNN for all zones and especially for lateral zones, which contained less amount of cartilage. Thus, these networks provided a more homogeneous segmentation accuracy across the 3D image. The example of the layer-to-layer performance of both U-Nets is illustrated in Fig.3 (for the case with an average performance of both networks). The averaged over the test dataset 3D DSC (0.77), as well as the averaged 2D DSC for medial slices (0.85), was the highest for the truncated U-Net. This CNN has also shown the best performance in terms of time (Fig. 2) - 0.05 s per slice. Further improvement may probably come from the transition of this architecture to 3D in order to utilize volumetric features of cartilage.Conclusions
In the current research, we have compared the performances of classical U-Net CNN and it’s truncated variant with a previously reported planar PB-CNN for segmentation of cartilage tissue in 3D VIBE MR images of wrists. U-Net architecture together with data augmentation provided an improved and homogeneous segmentation accuracy across 3D images. The truncated U-Net showed the best performance in terms of segmentation time and DSC values.Acknowledgements
This work was supported by the Russian Science Foundation (Grant No. 18-79-10167).References
[1] Ebrahimkhani S, et all. Artif Intell Med. 2020;106:101851
[2] Brui E, et al. NMR Biomed. 2020;33:e4320.
[3] Ronneberger O, et al. Med Image Comput Comput Assist Interv. 2015;9351:234-241.
Figures
Results for layer-to-layer analysis: average DSC values for the zones with different percentage of cartilage within the 3D images; average 3D DSC; average DSC for medial slices; the time needed for the segmentation of one slice. *The network was tested on a PC with common characteristics (an Intel Core i5-7640X processor with 32 Gb of RAM).