2876
Generating Cardiac Segmentation Masks of Real-Time Images from Self-Gated MRI to Train Neural Networks1Institute for Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2DZHK (German Centre for Cardiovascular Research), Göttingen, Germany, 3Campus Institute Data Science (CIDAS), University of Göttingen, Göttingen, Germany
Synopsis
Cardiac segmentation is essential for analyzing cardiac function. Manual labeling is relatively slow, so machine learning methods have been proposed to increase segmentation speed and precision. These methods typically rely on cine MR images and supervised learning. However, for real-time cardiac MRI, ground truth segmentations are difficult to obtain due to lower image quality compared to cine MRI. Here, we present a method to obtain ground truth segmentation for real-time images on the basis of self-gated MRI (SSA-FARY).
Introduction
Precise cardiac segmentation is an essential part for the analysis of cardiac function. Neural networks trained for the segmentation of the heart in short-axis view have achieved accuracies of over 90% for segmentation classes like left ventricle, right ventricle and myocardium, and have shown the potential of machine learning methods for fast, precise and reproducible computer-assisted diagnosis ¹. These algorithms are usually trained with supervised learning, which requires ground truth segmentations for each individual training image. The creation of such a dataset is especially difficult for images obtained with real-time cardiac MRI due to lower image quality and corresponding ambiguities of the tissue contours. We propose a method to automatically generate segmentation masks for real-time images by segmenting self-gated images obtained with SSA-FARY ². The segmentation of the self-gated images can be assigned to the real-time reconstructions by matching the corresponding cardiac and respiratory phases to the acquisition time of the data.Methods
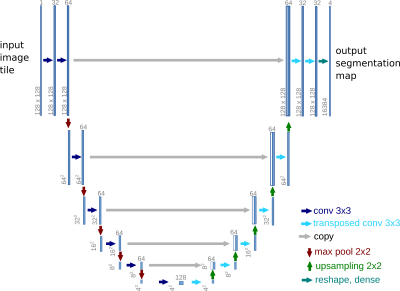
Cardiac images were acquired on a Siemens Skyra at 3T using a radial FLASH sequence (TE=3ms, TR=1.9ms) with a tiny golden angle of 7°, a flip angle of 10° and an in-plane resolution of 1x1 mm². Real-time reconstruction was performed using NLINV ³ and self-gated MRI was performed using SSA-FARY ² followed by a multi-dimensional reconstruction ⁴. All reconstruction steps were performed using BART ⁵.The self-gated images of one healthy volunteer were segmented with a U-Net-based neural network ⁶ implemented in BART (FIG_1) and trained with the Automated Cardiac Diagnosis Challenge (ACDC) ⁷ dataset, which features time series of cardiac short-axis views of 100 patients (4 pathology groups, 1 healthy group) and ground truth segmentations for left and right ventricular cavity and left ventricular myocardium. Images and segmentation masks have been augmented with spatial and brightness augmentations ⁸.
The self-gated images were cropped to center the heart and reduce computational costs of the training. For this purpose, the heart was localized with the Chan-Vese algorithm for contour detection ⁹ based on a method customized for cardiac MRI ¹⁰. The cropped, self-gated images were segmented by the network and the output was automatically post-processed to remove unsatisfying results. Segmentation results for the right ventricle were generally not accurate enough, so we focused on the results of left ventricle and myocardium. Pixel artifacts were excluded and holes of segmented structures were filled by computing convex hulls of ventricle and myocardium masks. After these post-processing steps, a segmentation of an image was judged as successful if the myocardium segmentation describes a full circle and no segmentation of the left ventricle is in contact with background pixels. The quality of the segmentation masks was evaluated by visual inspection.
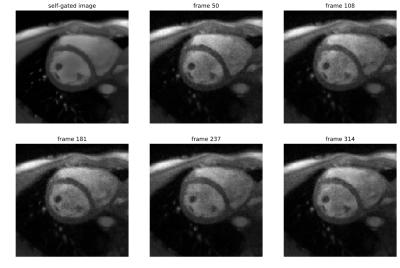
Under-sampled radial k-space data with 15 spokes per frame was used for real-time imaging. Real-time images were then matched to images obtained by self-gating, where multiple real-time images correspond to a single self-gated image (FIG_2). To minimize deviations, only the end-expiration state was used for further analysis. In this state, the spatial heart movement is minimal due to the consistently low lung volume. Due to a decreased lung volume, end-expiration can be identified as the respiratory state with the highest average image intensity in vicinity of the localized heart. The segmentation masks of the self-gated images were assigned to the matched real time images to obtain new training data.
Thereby generated training data was then used to re-train the network. The network was once further trained with noise-augmented ACDC training data and once with generated training data from real-time images. These two networks were then compared regarding the segmentation of previously unknown real-time images of another healthy volunteer for slices where cardiac segmentation is expected.
Results
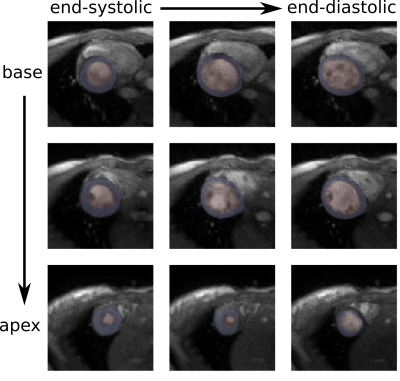
For end-expiration, we observe only minimal deviations between the segmentation mask and the structures in the real-time images (FIG_3).In a preliminary test, the network produces a higher amount of successful segmentations (myocardium segmentation describes a full circle) on a dataset of real-time reconstructed images if it has been re-trained with the generated segmentations and corresponding real-time images rather than with noise-augmented ACDC training data. Without re-training, 75% of images are successfully segmented. After re-training with eight epochs of generated training data from one healthy volunteer, 98% of images are successfully segmented. In similar training time, re-training with noise-augmented images showed no increase of the success rate.
Discussion and Conclusion
We presented a fast and easily accessible method to generate training data for cardiac segmentations of real-time images for supervised learning. The method allows to generate training data with an arbitrary amount of under-sampling while providing good segmentation mask. In contrast to mere noise-augmentation, this approach is more realistic as training images with under-sampling artifacts could be generated. However, the method needs further analysis.Acknowledgements
We were supported by the DZHK (German Centre for Cardiovascular Research) and funded in part by NIH under grant U24EB029240. We acknowledge funding by the "Niedersächsisches Vorab" initiative of the Volkswagen Foundation.References
[1] F. Isensee, et al. "Automatic Cardiac Disease Assessment on cine-MRI via Time-Series Segmentation and Domain Specific Features" STACOM 2017, LNCS, vol. 10663, pp. 120-129, March 2018.
[2] S. Rosenzweig, et al. "Cardiac and Respiratory Self-Gating in Radial MRI Using an Adapted Singular Spectrum Analysis (SSA-FARY)," IEEE Trans. Med. Imag., vol. 39, no. 10, pp. 3029-3041, Oct. 2020.
[3] S. Rosenzweig, et al. "Simultaneous multi‐slice MRI using cartesian and radial FLASH and regularized nonlinear inversion: SMS‐NLINV" Magn. Reson. Med., vol. 79, pp. 2057-2066, 2018.
[4] L. Feng et al., “XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing,” Magn. Reson. Med., vol. 75, no. 2, pp. 775–788, 2016.
[5] M. Uecker et al. "Berkeley advanced reconstruction toolbox." In Proc. Intl. Soc. Mag. Reson. Med., vol. 23, p. 2486, 2015.
[6] O. Ronneberger, P. Fischer, T. Brox. "U-Net: Convolutional Networks for Biomedical Image Segmentation" MICCAI, LNCS, vol. 9351, pp. 234-241, 2015.
[7] O. Bernard, A. Lalande, C. Zotti, F. Cervenansky, et al. "Deep Learning Techniques for Automatic MRI Cardiac Multi-structures Segmentation and Diagnosis: Is the Problem Solved?" IEEE Trans. Med. Imag., vol. 37, no. 11, pp. 2514-2525, Nov. 2018.
[8] F. Isensee et al. "batchgenerators -- a python framework for data augmentation" doi:10.5281/zenodo.3632567, 2020.
[9] T. Chan, L. Vese. "An Active Contour Model without Edges" Scale-Space Theories in Computer Vision, 1999.
10] G. Ilias, G. Tziritas. "Fast Fully-Automatic Cardiac Segmentation in MRI Using MRF Model Optimization, Substructures Tracking and B-Spline Smoothing" STACOM 2017, ACDC and MMWHS Challenges, pp. 91-100, 2018.
Figures