2871
Deep-learning cardiac MRI for quantitative assessment of ventricular volumes1Computer Science, Stony Brook University, Stony Brook, NY, United States, 2St. Francis Hospital, DeMatteis Center for Cardiac Research and Education, Greenville, NY, United States
Synopsis
The presented work introduces a deep-learning cardiac MRI approach to quantitative assessment of ventricular volumes from raw MRI data without image reconstruction. As the information required for volumetric measurements is less than that for image reconstruction, ventricular function may be assessed with less MRI data than conventional image-based methods. This offers the potential to improve temporal resolution for quantitatively imaging cardiac function.
Introduction
Quantitative assessment of ventricular volumes is essential for cardiac diagnostics. The clinical standard for volumetric measurements in cardiac MRI relies on the segmentation of cardiac images reconstructed from raw data1. The presented work is to develop a deep-learning approach to measuring ventricular volumes in the left ventricle (LV) and right ventricle (RV) directly from raw data without image reconstruction. Because the information required for volumetric measurements is less than that for image reconstruction, deep-learning cardiac MRI may be run with less raw data, providing an imaging speed gain over conventional image-based techniques.Methods
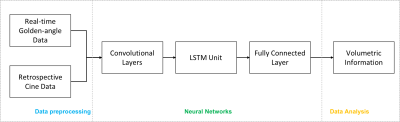
A convolutional neural network (CNN) model (Figure 1) was developed with 5 convolutional layers, a Long Short-Term Memory (LSTM)2 unit and a fully-connected layer. This model was trained to measure LV and RV volumes from k-space data collected in short-axis 2D cardiac MRI. The convolutional layers were used to extract k-space data features. The LSTM unit was used to process temporal correlation. The fully-connected layer was used to generate a final time series of volumetric measurements.The data for training and validation were collected with ECG-gated retrospective cine and real-time imaging. The retrospective cine was run using a balanced steady state free precession (bSSFP) sequence with the following parameters: FOV=340×(220-250)mm, voxel size=1.5-1.9mm, segments=5-8, iPAT factor=2, ECG-synchronized phases=30, TR/TE=2.6/1.3ms, FA=50°-75°, slice thickness=8mm, and bandwidth=1420Hz. The real-time imaging was run using a golden-angle radial bSSFP sequence with the following parameters: FOV=230-250mm, voxel size=1.5-1.9mm, TR/TE=2.2-3.0/1.1-1.5ms, FA=50°-75°, slice thickness=8mm, bandwidth=1510Hz, and 3072 golden-angle radial views.
The ground-truth volumetric measurements for training and validation were generated with a standard cardiac MRI approach: The retrospective cine and real-time images were reconstructed from raw data respectively1. A semi-automatic image segmentation method was used to measure ventricular volumes from the images3. The training was run with 237 datasets from retrospective cine imaging and 27 datasets from real-time imaging. The trained CNN model was validated with 20 datasets from retrospective cine and 4 datasets from real-time imaging. The training and validation were run on a NVIDIA V100 GPU with 16GB memory.
Results
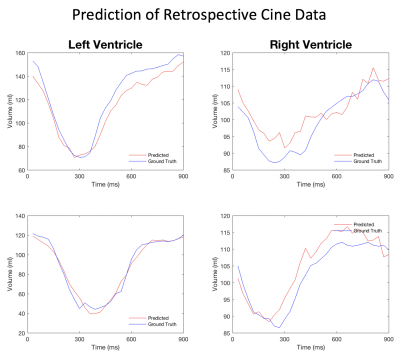
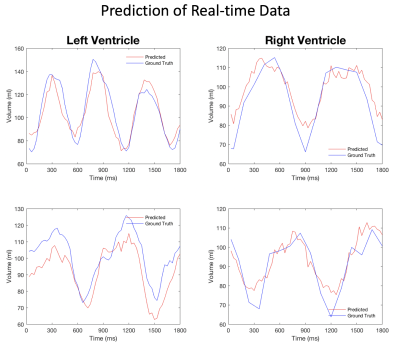
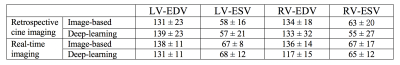
Figures 2 and 3 give a few examples from the validation experiments with retrospective cine and real-time golden-angle radial data. It can be seen that both measurements match the ground truth well. Table 1 lists all the measurements of end-diastolic and end-systolic volumes in the validation. They are comparable to the conventional image-based measurements. With deep-learning, however, every volume may be measured with 12 radial views or less. In comparison, image-based methods require at least 16 radial views for reconstructing an image with sufficient quality for image segmentation.Discussion
Deep-learning has been used to reconstruct images from raw data and perform segmentation in the reconstructed images. The presented work provides an approach to combining image reconstruction and segmentation into a single deep-learning model that can extract volumetric information directly from raw data. By skipping image reconstruction, ventricular volumes can be measured with less data. This imaging speed gain may be used to improve temporal resolution in volumetric measurements. In this work, we have found that deep-learning cardiac MRI has the potential to achieve a temporal resolution of 10-20ms in quantitative assessment of ventricular volumes. This will make possible to measure fast cardiac dynamics, e.g., isovolumetric contraction and relaxation.Conclusion
The presented work demonstrates a deep-learning model that can measure LV and RV volumes directly from k-space data in cardiac MRI. This deep-learning cardiac MRI method provides an imaging speed gain over conventional techniques that rely on image reconstruction and segmentation.Acknowledgements
None.References
1. Attili AK, Schuster A, Nagel E, Reiber JHC, van der Geest Rob J. Quantification in cardiac MRI: advances in image acquisition and processing. Int J Cardiovasc Imaging 2010; 26(Supplement 1): 27-49.
2. Hochreiter S, Schmidhuber J. Long short-term memory. Neural computation 1997; 9(8): 1735-1780.
3. Pluempitiwirlyawej C, Moura JMF, Wu Y-J, Lin HC, STACS: new active contour scheme for cardiac MR image segmentation. IEEE Trans Med Imag 2005; 24(5): 593-603.
Figures