2852
Time resynchronization of data obtained during cardiovascular MRI combined with catheterization using biophysical cardiac modeling1Inria, Palaiseau, France, 2LMS, Ecole Polytechnique, Palaiseau, France, 3Department of Cardiology, Boston Children’s Hospital, Boston, MA, United States, 4Division of Pediatric Cardiology, UT Southwestern Medical Center Dallas, Dallas, TX, United States, 5Pediatric department, Kafrelsheikh University, Kafr Elsheikh, Egypt, 6Department of Mathematics, Czech Technical University in Prague, Faculty of Nuclear Sciences and Physical Engineering, Prague, Czech Republic
Synopsis
Time resynchronization of data is often needed when the analysis is based on combining several cardiovascular MRI (CMR) sequences and/or when CMR is combined with a different modality such as pressure catheter during interventional CMR (iCMR). In the present work we propose using patient-specific biophysical model to resynchronize the intraventricular pressure and volume data. The model-driven resynchronization strategy generates pressure-volume (P-V) loops, which can be then used for clinical interpretation. Moreover, the patient-specific model provides additional mechanical indicators of patient's physiology. This framework has the perspective to contribute to planning of complex surgical interventions.
Introduction
The analysis of intraventricular pressure-volume (P-V) loop contributes to a detailed assessment of heart function, which can be of a significant help in planning a complex surgical intervention. Cardiovascular magnetic resonance imaging (CMR) combined with cardiac catheterization (invasive CMR, iCMR1) allows to record ventricular pressure during the MR acquisition. However, the recorded pressure and the volumes reconstructed from cine MRI are not precisely synchronous due to: (1) a limited temporal resolution of cine MRI (~30 ms used for iCMR); and since (2) the fluid-filled catheter is known to record the pressure changes with some delay, which varies depending e.g. on the catheter size or catheter softening due to blood temperature. Therefore, prior to plotting the P-V loop, the recorded intraventricular pressure and volumes need to be aligned in time.In this work we propose to create a patient-specific biophysical model, based on physiological assumptions, while using the information obtained from the iCMR data. This model is then used as a template for the time resynchronization of the P-V data. Additionally, to the “physical filtering” of the acquired P-V loop by the biophysical model, the patient-specific model can provide additional characteristics of the status of the patient’s cardiovascular system, which are not directly visible in the data and can contribute to planning the optimal clinical management.
Methods - Data
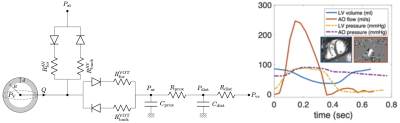
Data: Four patients were included in the present study – Patient #1 and 2 with repaired tetralogy of Fallot (rTOF) prior to pulmonary valve replacement, and Patients #3 and 4 with univentricular hearts (prior to completing the Fontan circulation). In rTOF patients, retrospective ECG gating was used (with parallel factor SENSE=2). In Fontan patients, due to a need of shortening the scanning time, a highly accelerated prospectively ECG-triggered sequence was used (kt-SENSE factor 6, partial Fourier 0.625). The target temporal resolution was ~30 ms in either case. The ventricular volumes were measured from the cine MRI using motion tracking algorithm2. The intraventricular pressures were recorded throughout the MR acquisition.Biophysical model of heart: The model of a single heart cavity3,4 was employed. The geometry and kinematics are reduced to a sphere with inner radius R and wall thickness d, while the constitutive mechanical laws are preserved as in full 3D heart model5. The circulation system is represented by a two-stage Windkessel model, which is coupled to the heart via system of diodes representing the cardiac valves (Figure 1).The passive properties of the model (myocardial stiffness) were adjusted according to the measured end-diastolic pressure and volume4. Maximum active stress developed by the model during the cardiac cycle (myocardial contractility) was adjusted according to the end-systolic volume. The myocardial stiffness and contractility represent examples of mechanical quantities accessed by the model and characterizing the physiology state of the heart.
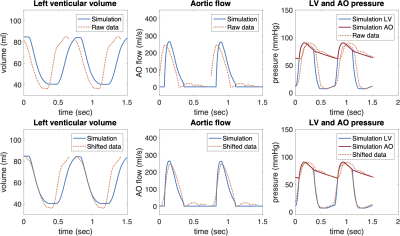
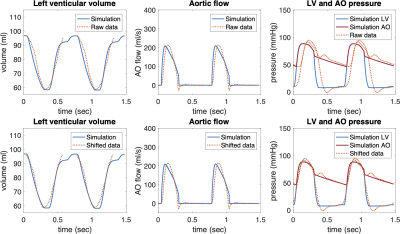
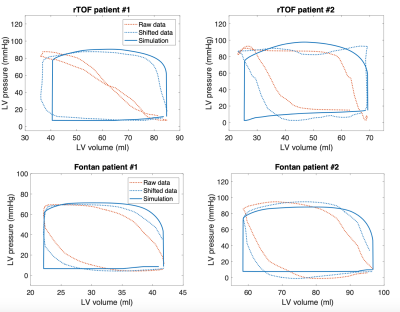
Time-resynchronization: Thanks to the biophysical and physiological character of the model, the timing of the cardiac phases (and volume and pressure changes) in the model was considered as a reference. The model was then used to time-resynchronize the acquired clinical data by minimizing the time offset between the acquired data and the reference model during cardiac cycle. P-V loops of original and resynchronized data were constructed.
Results
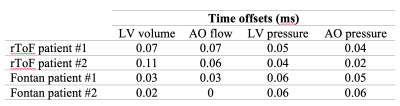
Figures 2 and 3 show an example of time-resynchronization of the data using biomechanical model for left ventricle (LV) of Patient #1 (rTOF, retrospective ECG gating) and Patient #3 (Fontan, prospective ECG triggering), respectively. Figure 4 shows the resynchronized P-V loops for all patients. Table 1 (in Figure 5) shows the time offsets obtained by the proposed procedure, which were then used to synchronize the data.Discussion
In this work we demonstrated that time-resynchronization by employing biomechanical model can be used to filter P-V loops in clinical settings both when retrospectively or highly accelerated prospectively ECG triggered cine sequence was used. Reconstruction of the full P-V loop whilst missing data in the early systole and late diastole (in prospective ECG triggering) exemplifies a model-based filtering of the iCMR data. The patient-specific parameters of the model, e.g. myocardial contractility and stiffness6, provide additional mechanical indicators of heart’s functional state. Presented modeling framework is fast and could be used in clinical settings, possibly even directly during the iCMR procedure.Conclusion
Patient-specific cardiac model is a promising time-resynchronization tool when combining several CMR sequences and/or when CMR is combined with catherization data.Acknowledgements
The study was supported by the Inria-UTSW Associated Team TOFMOD. R. Chabiniok additionally acknowledges the support of the Ministry of Health of the Czech Republic (project No. NV19-08-00071).
References
1. Veeram Reddy SR et al. Invasive cardiovascular magnetic resonance (iCMR) for diagnostic right and left heart catheterization using an MR-conditional guidewire and passive visualization in congenital heart disease. JCMR 2020;22(1):20.
2. Genet M et al. Equilibrated warping: Finite element image registration with finite strain equilibrium gap regularization. Med Image Anal. 2018;50:1-22.
3. Caruel M et al. Dimensional reductions of a cardiac model for effective validation and calibration. Biomech Model Mechanobiol. 2014;13(4):897-914.
4. Le Gall A et al. Monitoring of cardiovascular physiology augmented by a patient-specific biomechanical model during general anesthesia. A proof of concept study. PLOS ONE. 2020;15(5).
5. Chapelle D et al. An energy-preserving muscle tissue model: formulation and compatible discretizations. Int J Multiscale Comput Eng. 2012;10(2):189.
6. Ruijsink B et al. Dobutamine stress testing in patients with Fontan circulation augmented by biomechanical modeling. PLoS ONE, 15(2):e0229015, 2020.
Figures