2828
Myelin imaging derived quantitative parameter mapping compared to myelin water fraction
Yuki Kanazawa1, Masafumi Harada1, Yo Taniguchi2, Syun Kitano3, Nagomi Fukuda3, Yuki Matsumoto1, Hiroaki Hayashi4, Kosuke Ito2, Yoshitaka Bito2, and Akihiro Haga1
1Graduate School of Biomedical Sciences, Tokushima University, Tokushima, Japan, 2Healthcare Business Unit, Hitachi, Ltd., Tokyo, Japan, 3Faculty of Medicine, Tokushima University, Tokushima, Japan, 4Faculty of Health Sciences, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
1Graduate School of Biomedical Sciences, Tokushima University, Tokushima, Japan, 2Healthcare Business Unit, Hitachi, Ltd., Tokyo, Japan, 3Faculty of Medicine, Tokushima University, Tokushima, Japan, 4Faculty of Health Sciences, Institute of Medical, Pharmaceutical and Health Sciences, Kanazawa University, Kanazawa, Japan
Synopsis
We evaluated myelin imaging R1·R2* derived from QPM-relaxation parameters compared with the MWF derived from the same QPM gradient-echo dataset. Linear regression analysis indicated a correlation between R1·R2* products values and MWF values derived from same QPM dataset (R = 0.53, P < 0.0001).The R1·R2* products map derived from QPM offers as follow advantage; quantitative analysis; artifact free; and post-processing without fitting algorithm after QPM estimation. Myelin map derived from QPM can be applied for the quantitative assessment of white matter structure as substitute for MWF.
Introduction
Recently, reports of myelin visualization using magnetic resonance imaging (MRI) have been increasing. The interest in Myelin water fraction (MWF) imaging for detection of normal appearing white matter (NAWM) water content has been growing. Some derivation methods for MWF have been reported; e.g., multi-gradient-echo 1, gradient and spin-echo 2, T2 preparation imaging 3, multicomponent driven-equilibrium single-pulse observation of T1 and T2 (mcDESPOT) 4 and as well as others. However, there are often possibilities of numerical problems raising from curve fitting and T2 cut-off range settings. On the other hand, we developed novel Myelin imaging using physical parameters derived from quantitative parameter mapping (QPM) 5. The purpose of our study is to evaluate Myelin imaging derived from QPM-relaxation parameters compared with the MWF derived from the same QPM gradient-echo dataset.Materials & Methods
Figure 1 shows the preprocessing procedure for two-type myelin images, which was used to calculate the R1·R2* product and MWF from QPM dataset in this study. After On a 3 T MR system (Hitachi, Ltd.), QPM datasets of the brain (nine healthy volunteers; mean age, 41 years) were acquired with a three-dimensional radio frequency-spoiled steady state gradient-echo (3D-RSSG) sequence; the imaging parameters were echo times (TEs), 4.6-36.8 ms (∆TEs, 4.6 ms); repetition times (TRs), 10, 20, 32.1, and 41.3 ms; flip angles (FAs), 10 and 40 degrees; RF phases, 160, 162, 172, and 173. After acquiring QPM datasets, myelin maps using relaxation rate R1 and R2* based on the T1w/T2w ratio method as follows 5;$$\frac{T_{1}w}{T_{2}w}\approx\frac{\frac{1}{T_{1}}PD}{T_{2}^{*}PD}=\frac{R_{1}}{\frac{1}{R_{2}^{*}}}=R_{1}\cdot{R_{1}^{*}}$$
where T1, T2, and T2* are relaxation times and is proton density of a tissue. Signals of T1w and T2w images are derived from each relation as $$$T_1w\approx\frac{1}{T_1}PD,T_2w\approx T_2PD\approx T_2^*PD$$$. Using parameter map derived from QPM can be quantitatively visualized without signal bias. On the other hand, the MWF was derived using multi-component analysis based on T2* decay relaxometry of multi-gradient-echo sequence. The following bi-exponential fitting model (i.e., two-pool model) was used as follows;
$$S\left(t\right)=A_{short}\exp{\left(-\frac{TE}{T_{2,short}^*}\right)}+A_{long}\exp{\left(-\frac{TE}{T_{2,long}^*}\right)}$$
where S is the signal of each TE, A and T2* are amplitude and T2*relaxation time for short and long water pools, respectively. After fitting the measured decay signals, MWF is estimated as;
$$MWF=\frac{A_{short}}{A_{short}+A_{long}}$$
MWF was defined as the ratio of myelin water to total water was defined. MWF was drived from the same as QPM dataset, e.g., TEs = 4.6-36.8 ms; TR = 41.3 ms; FA = 40 degree. After setting regions of interest (ROIs) on bilateral WMs (e.g., frontal and occipital WM) and bilateral grey matter (GM) structures (e.g., caudate nucleus, putamen) in a basal ganglia slice for each subject, we measured the mean values and its standard deviation (SD). Measurements of R1·R2* products and MWF were compared for each region.
Results & Discussions
Figure 2 shows mean MWF and R1·R2* values of each region. Figure 3 shows representative histograms and maps of R1·R2* product and MWF obtained from QPM dataset. Figure 4 shows each myelin map of a representative subject. Figure 5 shows the relationship between R1·R2* products values and MWF values derived from same QPM dataset across all nine bilateral structures. Linear regression analysis shows a correlation between R1·R2* products values and MWF values derived from same QPM dataset (R = 0.53, P < 0.0001). The R1·R2* products map was observed with less field inhomogeneous area than the MWF map. The R1·R2* products map derived from QPM offers as follow advantage; quantitative analysis; artifact free; and post-processing without fitting algorithm after QPM estimation.Conclusion
Myelin map derived from QPM can be applied for the quantitative assessment of white matter structure as substitute for MWF.Acknowledgements
This study was partly supported by JSPS KAKENHI [grant number 20K07997].References
- Hwang D, Kim DH, Du YP. In vivo multi-slice mapping of myelin water content using T2* decay. Neuroimage 2010;52:198–204.
- Prasloski T, Rauscher A, MacKay AL, Hodgson M, Vavasour IM, Laule C, Mädler B. Rapid whole cerebrum myelin water imaging using a 3D GRASE sequence. Neuroimage 2012;63:533–539.
- Oh J, Han ET, Pelletier D, Nelson SJ. Measurement of in vivo multi- component T2 relaxation times for brain tissue using multi-slice T2 prep at 1.5 and 3 T. Magn Reson Imaging 2006;24:33–43.
- Lenz C, Klarhfer M, Scheffler K. Feasibility of in vivo myelin water imaging using 3D multigradient-echo pulse sequences. Magn Reson Med 2012;68:523–528.
- Kanazawa Y, Harada M, Taniguchi Y, et al. Myelin imaging derived from quantitative parameter mapping, Proc. ISMRM 2019;27:3313.
Figures
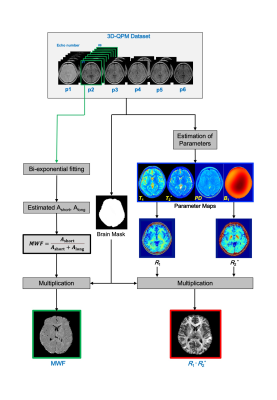
Fig.1 A schematic diagram of the QPM-myelin mapping (R1·R2*) and MWF images procedures.
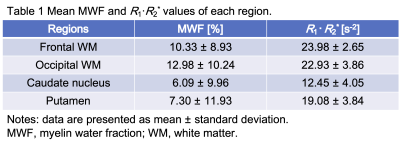
Fig. 2 Mean MWF and R1·R2* values of each region.
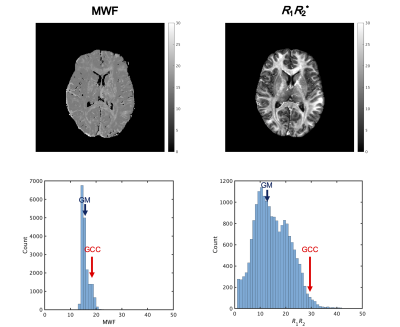
Fig. 3 Representative histograms and maps of R1·R2* product and MWF obtained from QPM dataset. Subject was 26-year-old man of healthy volunteer.
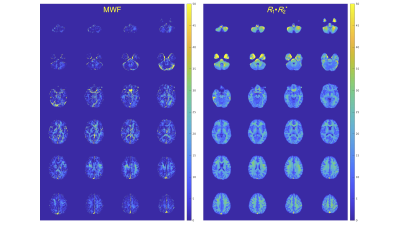
Fig. 4 Each myelin map of a representative subject (26-year-old man). Left shows MWF, and right R1·R2* map.
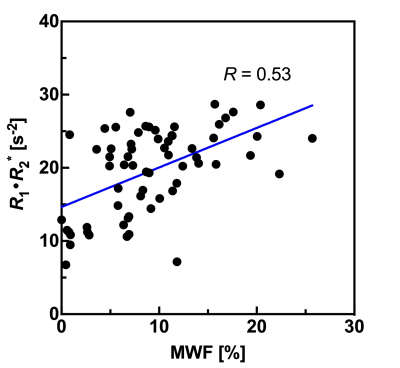
Fig. 5 Plots the relationship between MWF values and R1·R2* product values derived from QPM across all four bilateral structures.