2824
MS-Voter: Learning Where to Vote for Confluent Multiple Sclerosis Lesion Separation1Cornell University, New York, NY, United States
Synopsis
Lesion count, which encodes the lesion historical information, is an important biomarker for diagnosis and treatment of multiple sclerosis. Confluent lesions pose a great challenge to traditional automated methods, as these lesions are connected spatially, which requires expert experience to separate them. In this abstract, we propose a Hough voting method based on deep neural networks to resolve the issue. Experimental results on an in-house dataset demonstrates the superiority of our approach.
Summary
A machine learning technique is described for separating confluent lesions in multiple sclerosis using Hough voting and K-means clustering.Introduction
Lesion segmentation [1] is of vital importance for multiple sclerosis (MS) diagnosis and treatment, and many automated methods have achieved excellent performance based on recent advances in deep neural networks. However, lesion count cannot be obtained from the segmentation directly, as there exist many confluent lesions spatially connected with each other. Usually, these confluent lesions [2] can be separated by trained experts, but the process of which is tedious, time-consuming. Thus, in this abstract, we propose an approach called MS-voter to resolve the issue. In MS-voter, we use Hough voting [3] to determine the number of and center of confluent lesions in a large spatial connected lesion. With lesion centers, we further apply K-Means clustering to group voxels for confluent lesion separation. The advantage of the approach is that it considers both contextual information and lesion geometry.Methods
The MS-voter approach can be divided into four steps: 1) prepare ground-truth training data based on voxel offsets and voxel weights; 2) train the deep neural network using L1 loss; 3) perform network inference to get prediction of voxel offsets and voxel weights, followed by voting to determine lesions centers; 4) The lesion centers are used to initialize K-Means Clustering to get final lesion separation.Training Data Preparation:
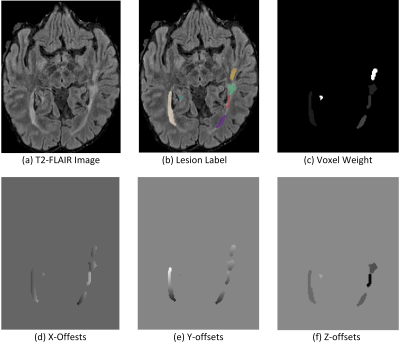
We use ground-truth lesion labels to generate voxel offsets and voxel weights. As can be seen from Figure 1, for each individual lesion, we first compute its mass center, and then use the coordinates of each voxel in that lesion to substracte the center coordinates to get voxel offsets to the center. Since the image is a 3D volume, we have three offsets, i.e. X-offsets, Y-Offsets, and Z-offsets. Besides, to train the network to vote for center, we also assign a voting weight to each voxel. We compute an average lesion size $$$\hat{S}$$$, and for voxel in each individual lesion with size$$$\hat{S}_i$$$, the voxel weight for that lesion is $$$\frac{\hat{S}}{S_i}$$$.
Network Training Using L1 Loss:
Once we get ground-truth voxel offsets and voxel weights, we can use a fully convolutional neural network such as U-Net to train the model. The input of the model is multi-sequence MRI data, T1-w, T2-w, and T2-FLAIR, and the output is the voxel offsets and voxel weights. We use L1 loss to train the network as follows:
$$L_1 = \frac{-1}{N}(|X-\hat{X}|+|Y-\hat{Y}|+|Z-\hat{Z}|+|W-\hat{W}|), $$
where $$$N$$$ is the total number of voxels, $$$X,Y,Z$$$ are ground-truth voxel offsets, $$$W$$$is the ground-truth voxel weight, and $$$\hat{X},\hat{Y},\hat{Z},\hat{W}$$$ are corresponding network predictions.
Network Inference With K-Means Clustering:
Once the network is trained, given a new image and segmentation mask, we can output corresponding voxel offsets and voxel weights. For each voxel, we can get its coordinates, x-offset, y-offset, z-offset, and voting weight. Suppose we have an empty 3D Hough voting map with the same size as the input image, and $$$\Omega$$$ is the spatial coordinate space of the image, and $$$v\in \Omega$$$ is an index vector indicateing where the voxel is in the image. Suppose the coordinate of a voxel is $$$v$$$, with offsets we can compute the center it points to as $$$\hat{v}=(\hat{X}_v,\hat{Y}_v,\hat{Z}_v)$$$. We then accumulate the weight in $$$H$$$ as $$$H_{\hat{v}}=H_{\hat{v}}+\hat{Z}_v$$$.
With the Hough voting map, we apply non-maximal suppression and thresholding to get the final binary map indicating lesion centers. We then use the coordinates of these centers to initialize K-Means clustering, for each spatially connected lesion, we run a separate K-Means with lesion center initializ€ation to get final labels of each lesion.
Results
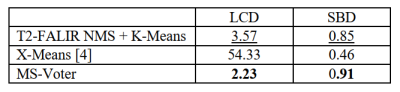
We use two metrics to evaluate our methods. Absolute lesion count difference (LCD) is used to measure the absolute difference of predicted lesion count and ground-truth lesion count. The symmetric best Dice (SBD) metric averages Dice score between pairs of predicted and the ground truth labels yielding maximum Dice. We first implemented two baseline methods for comparison as follows:T2-FALIR NMS + K-Means: Since lesions are hyper-intensive on T2-FLAIR image, we perform non-maximal suppression on T2-FLAIR image to select local maximal points [5] as potential lesion centers. Once we get the lesion centers, we perform K-Means clustering for lesion separation.
X-Means [4]: This method improves K-Means by determining the number of K automatically using information theory, and thus we can directly apply it for lesion separation.
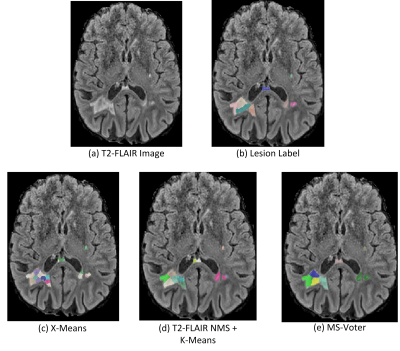
We can see from Figure 3 that X-Means performs worst, as it performs separation based geometric locations and ignores contextual information. T2-FALIR NMS + K-Means performs better than X-Means, as we provide additional lesion intensity information to guide the separation. Our proposed MS-Voter has demonstrated the superiority compared to the former methods, as our lesion centers are predicted by trained deep neural networks that can capture richer contextual information.
Discussion and Conclusion
In this abstract, we proposed a MS-Voter approach for confluent lesion separation. In MS-Voter, each voxel is trained to vote for the lesion center it belongs to. With deep neural networks trained on numerous data samples, compared to traditional methods our MS-Voter can capture richer contextual information as well as the lesion geometric property, resulting in a reasonably good performance.Acknowledgements
No conflict of interest.References
1. Zhang, Hang, Jinwei Zhang, Qihao Zhang, Jeremy Kim, Shun Zhang, Susan A. Gauthier, Pascal Spincemaille, Thanh D. Nguyen, Mert Sabuncu, and Yi Wang. "RSANet: Recurrent Slice-wise Attention Network for Multiple Sclerosis Lesion Segmentation." In International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 411-419. Springer, Cham, 2019.
2. Dworkin, Jordan D., Kristin A. Linn, Ipek Oguz, Greg M. Fleishman, Rohit Bakshi, Govind Nair, Peter A. Calabresi et al. "An automated statistical technique for counting distinct multiple sclerosis lesions." American Journal of Neuroradiology 39, no. 4 (2018): 626-633.
3. Illingworth, John, and Josef Kittler. "A survey of the Hough transform." Computer vision, graphics, and image processing 44, no. 1 (1988): 87-116.
4. Pelleg, Dan, and Andrew W. Moore. "X-means: Extending k-means with efficient estimation of the number of clusters." In Icml, vol. 1, pp. 727-734. 2000.
5. Zhou, Xingyi, Dequan Wang, and Philipp Krähenbühl. "Objects as points." arXiv preprint arXiv:1904.07850 (2019).
Figures