2789
A Video Block Matching 3D Transform-domain Collaborative Filtering Approach for High Spatiotemporal Resolution DCE-MRI
Zhongbiao Xu1, Zhenguo Yuan2, Yaohui Wang3, Junying Cheng4, Rongli Zhang5, Ling Xia6, Yanqiu Feng7, Feng Liu8, and Zhifeng Chen7
1Guangdong Provincial People's Hospital, guangzhou, China, 2shandong medical imaging research institute ,shandong provincial hospital ,afflliated shangdong first medical uinversity, jinan, China, 3Institute of Electrical Engineering, Chinese Academy of Sciences, Beijing, China, 4First Affiliated Hospital of Zhengzhou University, zhengzhou, China, 5Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hongkong, China, 6Department of Biomedical Engineering, Zhejiang University, HangZhou, China, 7Southern Medical Southern, guangzhou, China, 8School of Information Technology and Electrical Engineering, The University of Queensland, Brisbane, Australia
1Guangdong Provincial People's Hospital, guangzhou, China, 2shandong medical imaging research institute ,shandong provincial hospital ,afflliated shangdong first medical uinversity, jinan, China, 3Institute of Electrical Engineering, Chinese Academy of Sciences, Beijing, China, 4First Affiliated Hospital of Zhengzhou University, zhengzhou, China, 5Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hongkong, China, 6Department of Biomedical Engineering, Zhejiang University, HangZhou, China, 7Southern Medical Southern, guangzhou, China, 8School of Information Technology and Electrical Engineering, The University of Queensland, Brisbane, Australia
Synopsis
High spatiotemporal resolution DCE-MRI has great clinical value in disease diagnosis and treatment. In this study, we propose to use a video block matching 3-D filtering approach to improve high spatiotemporal resolution DCE-MRI. Both phantom and in vivo experiments were performed in this work. The phantom experiment indicated that the proposed approach outperforms iGRASP and GRASP-Pro methods with lower reconstruction errors, especially in cases involving super high reduction factors. In vivo experiments draw similar conclusion. This new technique can provide a potential solution for real-time imaging and image guided radiation therapy.
Purpose
DCE-MRI is widely used in perfusion imaging [1]. High spatial and temporal resolutions are both the pursue of ideal DCE-MRI [2]. However, there is a tradeoff between spatial and temporal resolution. When reduction factor goes higher, image quality decreases quickly due to noise amplification and artefacts remaining. BM3D block matching is an excellent denoising method for handling white Gaussian noise, which is particularly appropriate for processing MR images [3]. In this study, we propose to use a video block matching sparse 3D transform-domain collaborative filtering (vBM3d) to reconstruct DCE-MRI data [4]. Golden-angle radial acquisition strategy is getting popular in DCE-MRI due to its flexible reconstruction strategy and high SNR benefit [5–9]. We use the golden-angle radial acquisition to verify the proposed approach. Phantom and in vivo experiments were both performed in this study, and demonstrated that the proposed scheme can generate higher PSNR, lower RMSE and better structure similarity (SSIM) in the cardiac DCE-MRI phantom experiments, especially in high under-sampling cases. For in vivo experiments, compared to iGRASP and GRASP-Pro, the proposed method provides higher SNR without diagnostic information loss. The scores of the radiologist also proved the conclusion.Method & Experiments
VBM3d takes the redundancy information of one frame as well as other dynamic frames for denoising [4]. It is assumed that the standard deviation of the noise is known in advance. We incorporate the vBM3d technique into the reconstruction procedure and iteratively solved the formed inverse problem.A cardiac DCE-MRI simulation data was generated using MRXCAT phantom [10]. The basic simulation parameters were as follows: reconstruction matrix 192×192, 32 dynamic time frames, number of channels 12, and random noise are added in both real and imaginary components to yield an SNR of 30 dB. This phantom dataset was then inversely gridded into golden-angle radial trajectory frame-by-frame using NUFFT toolbox [11]. Ground truth data were pre-generated for numerical evaluation.
An in vivo liver DCE-MRI experiment was performed on a 3.0 T Vida MR scanner (Siemens AG Medical Solutions, Erlangen, Germany) using a 24-channel body/spine coil array. A radial 3D stack-of-stars FLASH pulse sequence with free-breathing golden-angle sampling scheme was performed for this imaging test. The relevant parameters were set as the following: FOV=380 × 380 × 240 mm3, TR/TE = 3.6/1.55 ms, number of slices 44, number of readout points in each spoke 512, oversampling ratio 2, number of spokes 1913, and slice thickness 5 mm, spatial resolution 1.5x1.5x5 mm3. Another liver DCE-MRI dataset was downloaded from https://cai2r.net/resources/software/grasp-matlab-code. In this work, all computations were implemented in Matlab (R2015b; the Mathworks, Natick, MA, USA), for off-line reconstruction on a Linux server (Red Hat Enterprise, Core i7 Intel Xeon 2.8 GHz CPUs and 64GB RAM).
Data Analysis
In this study, RMSE, PSNR and SSIM index[12] are used for the evaluation of cardiac phantom data. In order to globally evaluate the DCE-MRI dataset, we choose the averaged value among all time frames of each criteria for all the comparison methods.To evaluate the image quality of one in vivo dataset of different reconstruction approaches, a radiologist with 17 years’ experience on abdominal imaging scored all the images using a 5-level score protocol. Then the scores were analyzed with ANOVA using Excel (Microsoft, Redmond, WA, USA), P < 0.05 was considered statistically significant.
Results & Discussion
We validated the proposed approach by imaging a cardiac DCE-MRI phantom, the results were shown in Fig 1 and Table 1. From the table, it can be clearly seen that the proposed vBM3d scheme outperformed the CG-SENSE, iGRASP and GRASP-Pro methods in the cardiac perfusion phantom test. All the approaches were optimized based on RMSE. It is found that vBM3d results in higher PSNR and SSIM, and it preserves image details better than other compared approaches. This were especially obvious in cases with high reduction factors.For in vivo experiments, the denoising effect (Figs 2 & 3) can be seen. It is obviously seen that the vBM3d produced cleaner images than other compared approaches. The reconstructed images in Fig. 3 were scored and the statistical results were demonstrated in Fig. 4. For score comparison of all the tested approaches, vBM3D>GRASP-Pro>iGRASP>CG-SENSE. The proposed method generated significantly better results than iGRASP (P=0.01), also GRASP-Pro can have significantly better image quality than GROWL (P < 0.05). This was previously proved by Ref.[9]. When comparing vBM3d with GRASP-Pro, although the mean score of vBM3d is slightly higher than GRASP-Pro, the p value suggested that there was no significant difference between these two methods. Hence, the proposed scheme can generate comparable results to GRASP-Pro.
Due to the advantage of proposed vBM3d in experiments with high reduction factors, it has clinical potential of real-time imaging. Further studies will focus on the investigation of denoising effect of dynamic MRI with different noise levels.
Conclusion
Our proposed vBM3d approach is a video block matching approach for high spatiotemporal DCE-MRI. Compared to current iGRASP-type techniques, it offers better image quality, including higher PSNR and SSIM with lower RMSE. Due to denoising effect and SNR benefit,it has a latent advantage of better temporal resolution, which can improve the potential of clinical DCE-MRI in real-time imaging and image guided radiation therapy.Acknowledgements
This work was supported by NSFC grant (61801205), CPSF grant (2018M633073), OCPC fellowship and Guangdong Basic and Applied Basic Research Foundation (2019A1515111182).References
[1] Aronhime S, Calcagno C, Jajamovich GH, et al. DCE-MRI of the liver: effect of linear and nonlinear conversions on hepatic perfusion quantification and reproducibility. J Magn Reson Imaging. 2014; 40: 90–98. [2] Pari V. Pandharipande, Glenn A. Krinsky, Henry Rusinek, et al. Perfusion Imaging of the Liver: Current Challenges and Future Goals. 2005; 234: 661–673. [3] K. Dabov, A. Foi, V. Katkovnik, et al. Image denoising with block-matching and 3D filtering. Proc. SPIE Electronic Imaging '06, no. 6064A-30, San Jose, California, USA, January 2006. [4] K. Dabov, A. Foi, K. Egiazarian. Video denoising by sparse 3D transform-domain collaborative filtering. European Signal Processing Conference (EUSIPCO-2007), September 2007. [5] Winkelmann S, Schaeffter T, Koehler T, et al. An optimal radial profile order based on the golden ratio for time-resolved MRI. IEEE Trans Med Imaging. 2007; 26: 68–76. [6] Feng L, Grimm R, Block KT, et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn Reson Med. 2014; 72: 707–717. [7] Chen Z, Kang L, Xia L, et al. Sequential combination of parallel imaging and dynamic artificial sparsity framework for rapid free-breathing golden-angle radial dynamic MRI: K-T ARTS-GROWL. Med Phys. 2018; 45: 202–213. [8] Feng L, Axel L, Chandarana H, et al. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med. 2016; 75: 775–788. [9] Feng L, Wen Q, Huang C, et al. GRASP-Pro: imProving GRASP DCE-MRI through self-calibrating subspace-modeling and contrast phase automation. Magn Reson Med. 2020; 83: 94–108. [10] Wissmann L, Santelli C, Segars WP, et al. MRXCAT: realistic numerical phantoms for cardiovascular magnetic resonance. J Cardiovasc Magn R. 2014;16:2–11. [11] https://cims.nyu.edu/cmcl/nufft/nufft.html. [12] Wang Z, Bovik AC, Sheikh HR, et al. Image quality assessment: from error visibility to structural similarity. IEEE T Image Process. 2004; 13: 600–612.Figures
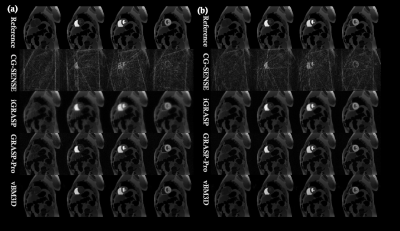
FIG.1. Comparison of the reconstruction
results of cardiac DCE-MRI phantom. (a) 38x under-sampling
experiment (5-spoke/frame). (b) 24x under-sampling experiment
(8-spoke/frame).
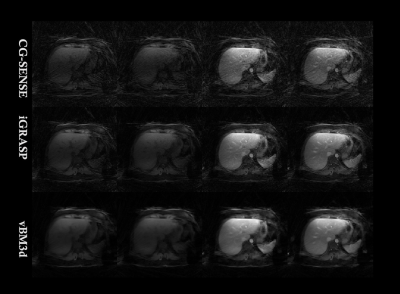
FIG. 2. Reconstruction images comparison
among different approaches for the first liver DCE-MRI dataset.
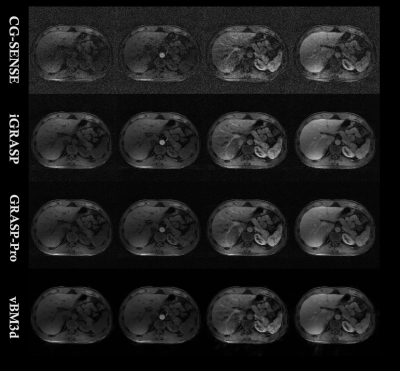
FIG.3. The reconstruction results of CG-SENSE (first row), iGRASP
(second row), GRASP-Pro (third row) and the proposed vBM3d framework (last row)
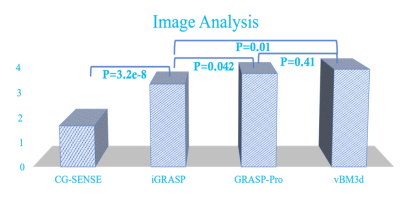
FIG.4. The score comparison of CG-SENSE,
iGRASP, GRASP-Pro and vBM3d.
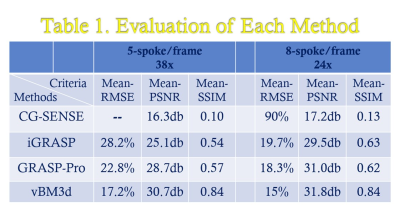
TABLE 1. The evaluation criterion for dynamic cardiac phantom