2760
Voxel-wise hepatocellular function prediction using population-based probability density function
Monchai Phonlakrai1, Behzad Asadi2, Neda Gholizadeh3, Kate Skehan2, Liam Hilleary2, Jameen Arms4, Saadallah Ramadan5,6, John Simpson2,3, Jonathan Goodwin2,3, Jarad Martin2,7, Yuvnik Trada2, Swetha Sridharan2,7, and Peter Greer2,3
1School of Health Sciences, The University of Newcastle, Newcastle, Australia, 2Radiation Oncology, Calvary Mater Newcastle Hospital, Newcastle, Australia, 3School of Mathematical and Physical Sciences, The University of Newcastle, Newcastle, Australia, 4Diagnostic Radiology, Calvary Mater Newcastle Hospital, Newcastle, Australia, 5Faculty of Health and Medicine, The University of Newcastle, Newcastle, Australia, 6HMRI Imaging Centre, John Hunter Hospital, Newcastle, Australia, 7School of Medicine and Public Health, The University of Newcastle, Newcastle, Australia
1School of Health Sciences, The University of Newcastle, Newcastle, Australia, 2Radiation Oncology, Calvary Mater Newcastle Hospital, Newcastle, Australia, 3School of Mathematical and Physical Sciences, The University of Newcastle, Newcastle, Australia, 4Diagnostic Radiology, Calvary Mater Newcastle Hospital, Newcastle, Australia, 5Faculty of Health and Medicine, The University of Newcastle, Newcastle, Australia, 6HMRI Imaging Centre, John Hunter Hospital, Newcastle, Australia, 7School of Medicine and Public Health, The University of Newcastle, Newcastle, Australia
Synopsis
Dynamic gadoxetate contrast-enhanced MRI yields spatial hepatocellular function through hepatic extraction fraction map. This allows well-functioning hepatocyte sparing in radiotherapy to avoid radiation-induced liver toxicity. However, the major challenge of using this parametric map in a clinical practice for normal function sparing is the lack of standard method to determine liver function at a voxel level within the same patient. As such, population-based kernel density function was proposed to deal with this problem to predict voxel-based probability of liver function. This novel approach also allows derivation of functional probability map that could be used for radiation beam guidance in function-based radiation treatment planning.
Introduction
Hepatic extraction function (HEF) map, derived from gadoxetate dynamic contrast-enhanced (DCE) MRI, allows focal liver function assessment and quantification [1]. However, use of HEF is still limited in a routine clinical setting, particularly, function-based radiation treatment planning. This is due to lack of a standard method that can identify high and poor functioning voxels in the same patient for clinical sparing. Therefore, this study proposes a novel approach for predicting voxel-wise liver function in HEF map using a population-based probability density function. This will ultimately facilitate function-based radiation treatment planning.Methods
SubjectsWe collected sixty-four clinical gadoxetate DCE-MRI, consisting of 25 normal liver function patients and 39 hepatocellular carcinoma (HCC) patients with underlying cirrhotic liver from Calvary Mater Newcastle Hospital, Australia. Normal liver function patients were identified from biochemical blood tests, history of non-chronic liver disease, and negative MRI study. The study was approved by the local and regional regulatory ethics committee (reference number 2019/ETH13629).
Hepatic extraction fraction (HEF) map generation
A voxel-wise HEF map was derived from clinical datasets of six-phase gadoxetate DCE-MRI, performed with 3T MRI. The method is based on truncated singular value decomposition as a deconvolution analysis technique, described in [2] and [3] with minor modifications. First, we applied least-square curve fitting to smooth original time-intensity curves of portal vein and liver using a shape-preserving piecewise cubic Hermite interpolating polynomial (pchip) method. The fitted values of the curves with 15 data-point intervals over the length of imaging time were extracted and used to compute HEF values in the model. Second, the hepatic retention phase was shortened to 15 min after injection to match the clinical dataset, usually lasted 15-20 min after injection.
Population-based probability density estimations
We used 62 out of 64 patients to generate the probability density function (PDF) of HEF values, and the remaining 2, consisting of one normal and one HCC patient, for the prediction. The PDF of HEF values in normal and HCC patients with cirrhotic liver were generated, separately. This was performed using the kernel smoothing function of Matlab according to the following equation
$$p(x)=\frac{1}{n}\sum_{i=1}^nK_w (x- x_i ), $$
where $$$p(x)$$$ is the probability density function, $$$x_i, i=1,2,...,n,$$$ are samples from an unknown distribution, $$$n$$$ is the sample size, $$$K(·)$$$ is the kernel smoothing function, and $$$w$$$ is the bandwidth.
Voxel-wise liver function prediction using population-based probability density function
The HEF maps from one normal liver function patient and one HCC patient were used for predicting the probability of liver function at the voxel level using following equation.
$$ρ(hef_{i,j}) = \frac{p(hef_{i,j})}{p(hef_{i,j})+p'(hef_{i,j})},$$
where $$$hef_{i,j}$$$ is the HEF value at pixel $$$(i,j)$$$, $$$p(hef_{i,j})$$$ is the PDF of HEF value for the normal-liver-function population, $$$ p'(hef_{i,j})$$$ is the PDF of HEF value for HCC population and $$$ρ(hef_{i,j})$$$ is the predicted probability of liver function for a given HEF value.
Data analysis
We calculated the mean, standard deviation, and coefficient of variation (CV) for both the HEF values and the predicted liver function probabilities in a normal liver function patient and a HCC patient with underlying cirrhotic liver. The CV was considered as excellent for less than or equal to 10%, good for between 10–20%, acceptable for between 20–30%, and poor for greater than 30%.
Results
This study demonstrates the successful predicting of hepatocellular function at the voxel level using the population PDF of HEF map. The PDFs are illustrated in Figure 1. The results of voxel-wise liver function prediction are shown in Figure 2 where the probability maps of a normal liver function patient and a HCC patient are provided as a comparison. In the normal liver function patient and the HCC-patient with underlying cirrhotic liver, the mean HEF values were 0.27± 0.05 and 0.14± 0.07, the mean predicted probability values were 0.80±0.17 and 0.33±0.27, and the CV of functional probability across voxels in probability maps were 21.05 % and 83.89 %, respectively.discussion
In this study, we have proposed a flexible method to predict liver function at a voxel level without labeling or training the data. The homogenous probability values of normal liver function patient was illustrated as expected (CV < 30%) as we hypothesized that hepatocyte function should be homogenous in a normal liver function patient. In contrast, the heterogeneity of probability values in HCC patient with underlying liver cirrhosis was observed (CV > 30%) in comparison with the normal liver function patient. This is probably caused by the destruction of normal architecture and the loss of hepatocytes in cirrhotic patients [4].Conclusion
This study explored the usage of population-based PDF of HEF values for voxel-wise hepatocellular function prediction. The results also show the potentials in the clinical use of this method for well-functioning hepatocyte sparing in function-based radiation treatment planning.Acknowledgements
We would like to express our gratitude to the following individuals for their expertise and assistance: Jose Baozi Ortega (Medical Physic research group, School of Mathematics and Physical Science, The University of Newcastle, Australia). Monchai Phonlakrai gratefully acknowledges the funding received towards his Ph.D. from the Chulabhorn Royal Academy, Thailand.References
- Nilsson H, Blomqvist L, Douglas L, Nordell A, Jonas E. Assessment of liver function in primary biliary cirrhosis using Gd-EOB-DTPA-enhanced liver MRI. HPB. 2010;12(8):567-76. doi:10.1111/j.1477-2574.2010.00223.x
- Ryeom H-K, Kim S-H, Kim J-Y, Kim H-J, Lee J-M, Chang Y-M, et al. Quantitative evaluation of liver function with MRI Using Gd-EOB-DTPA. Korean J Radiol. 2004;5(4):231-9. doi:10.3348/kjr.2004.5.4.231
- Nilsson H, Nordell A, Vargas R, Douglas L, Jonas E, Blomqvist L. Assessment of hepatic extraction fraction and input relative blood flow using dynamic hepatocyte‐specific contrast‐enhanced MRI. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2009;29(6):1323-31. doi:/10.1002/jmri.21801
- Ferrell L. Liver Pathology: Cirrhosis, Hepatitis, and Primary Liver Tumors. Update and Diagnostic Problems. Modern Pathology. 2000;13(6):679-704. doi:10.1038/modpathol.3880119
Figures
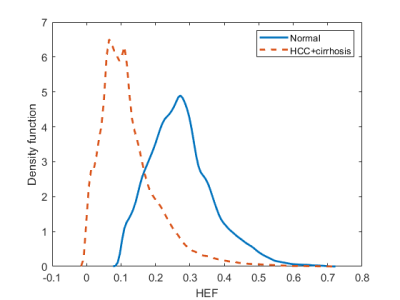
Figure1. The probability density function of normal liver function (n=24) and HCC with underlying liver cirrhosis (n = 38) populations were generated using kernel smoothing function.
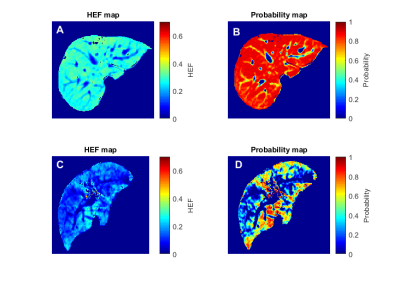
Figure2. The figure illustrates the comparison of HEF map (A, C) and the probability of liver function maps(B, D) between a normal liver function patient (upper row) and an HCC patient with underlying cirrhosis (Child-Pugh A) in the bottom row.