2730
BATS: the Boston ASL Template and Simulator – development and initial evaluation1Division of MRI research, Department of Radiology, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, MA, United States
Synopsis
Neuroscientific and clinical research has seen tremendous advances over the past 25 years. While hardware, pulse-sequences and reconstruction developments have been an immense driver for this, image processing tools have also been crucial.While anatomical templates such as the MNI152 have become widely used and are a hallmark of neuroimaging analyses, functional contrast-specific templates for group studies are much scarcer but of great interest. We therefore report here the initial development of BATS – the Boston ASL Template and Simulator, highlighting its development and initial use.
Introduction
Neuroscientific and clinical research has seen tremendous advances over the past 25 years. While hardware, pulse-sequences and reconstruction developments have been an immense driver for this, image processing tools have also been crucial. Among those, the possibility of spatially normalizing large datasets enabled group studies of brain structure and function and their modifications in altered states due to disease or physiological processes such as aging1. Spatial normalization usually requires a template in a normalized space to register data to. While anatomical templates such as the MNI152 have become widely used2,3 and are a hallmark of neuroimaging analyses such as VBM4, functional contrast-specific templates for group studies are much scarcer but of great interest5. For that matter, while a pediatric brain perfusion template exists6, adult average ASL perfusion templates that combine perfusion-weighted with contrast flexibility, blood-flow, transit-time are missing. We therefore report here the initial development of BATS – the Boston ASL Template and Simulator: an ASL-based average adult perfusion template based on high-resolution 3D variable-density FSE volumes, associated with anatomical contrasts for ease of use in conjunction with already existing tools, as well as a synthetic ASL image simulator allowing the creation of various averages with different perfusion contrasts as desired.Material and methods
Data collection: Data from 10 healthy volunteers were used to build the template. These subjects were scanned at 3T (GE Discovery MR750) using a 32-channel head coil, using 1mm3 T1-weighted MP2RAGE (for T1-weighted and T1-mapping) and T2-FLAIR (using a VFA 3D-FSE sequence), as well as low-resolution sequential multi-delay ASL scans to derive ATT, and high-resolution (1.6mm)3 whole brain ASL data (2.2/1.8s labeling/PLD) using a variable-density FSE sequence and Compressed-Sensing reconstruction. Details on the acquisition protocol have been previously reported7.Template construction: We used the Advanced Normalization Tools (ANTs) package for template construction8,9. Using the iterative multivariate template building framework, we chose 3 contrasts for template construction (FLAIR, ASL and T1-weighted) and used the following parameters: 5 outer iterations, B-Spline regularized SyN transform10 for the non-linear co-registration, cross-correlation metric, gradient step =0.0005, CPU parallelization. All data were initially co-registered with a rigid followed by affine registration prior to non-linear deformations to the average. All the template construction steps were performed at the ASL native resolution (1.6mm isotropic), building symmetric templates that were formed by mirroring individual images. Finally, the T1-weighted template built was non-linearly registered to the MNI152 1-mm template to allow benefiting from having the BATS template in a normalized space. To highlight this, we then computed CBF values in ROIs derived from the Harvard-Oxford subcortical atlas11.
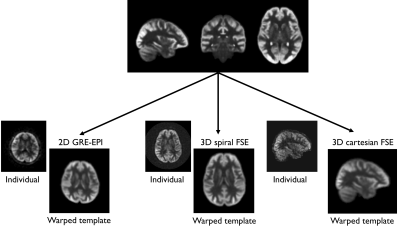
Evaluation:We evaluated the benefits of an ASL template using 3 individual ASL datasets acquired on the same 3T system using two different 32-ch receiver coils using either a 2D, unsuppressed axial gradient-echo EPI with simultaneous multi-slice (SMS) excitation (1.5/1.8s labeling/PLD, 30 pairs, 64x64x32 matrix, 3.5x3.75x4mm3), a 3D background-suppressed Stack-of-Spirals FSE (axial orientation, 1.5/2s labeling/PLD, regridded to a 128x128x36 matrix, reconstructed resolution 1.9x1.9x4mm3) and a 3D sagittal background-suppressed Cartesian FSE (2.2/1.8s labeling/PLD, 96x96x58, 2.3x2.3x3mm3). We then registered the BATS perfusion template to each dataset using a combination of affine and non-linear registration using ANTs (cross-correlation metric).
ASL simulator:Thanks to the availability of all required parameters, we used a standard kinetic model12,13 assuming two-compartments to be able to simulate synthetic ASL contrasts based on user-defined imaging parameters (labeling duration, PLD …) using the following equation:
$$dM=2\times M^0_t\times\alpha\times T^t_1\times f\times\exp(-\frac{\delta}{T_{1,b}})\times (\exp(-\frac{\max(w-\delta,0)}{T_{1,t}})-\exp(-\frac{-\max(\tau+w-\delta,0)}{T_{1,t}}))/\lambda$$
Results and Discussion
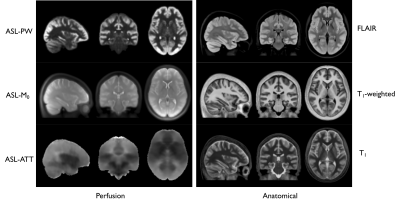
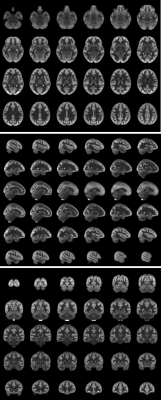
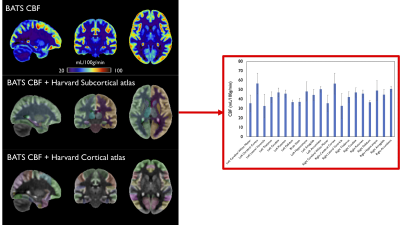
Fig1 shows the different components of the BATS template (perfusion-weighted, M0, ATT), as well as simultaneously provided T1-w/T1 and FLAIR averages that are readily in the MNI space. Reformats in the three planes are shown in Fig2 showing the high level of anatomical details provided by the BATS perfusion template especially when looking at the cortical gray matter. Several interesting features are observable thanks the power of group averaging, such as extremely strong signal originating from the choroid plexuses but also high perfusion signal and CBF for example in the primary motor areas.The availability of different required parameters allowed computing CBF templates that use either a single-compartment model or two-compartment models correcting for ATT and potentially T1. Fig 3 shows of the Harvard-Oxford cortical and subcortical atlases overlaid on the BATS CBF template, as well as quantification in subcortical ROIs.
When registering the template to individual scans acquired with various sequences and orientations, it was found to be fast and accurate as seen in Fig4 although a more detailed quantitative analysis of registration accuracy compared to standard registration frameworks using an intermediate anatomical scans should be done.
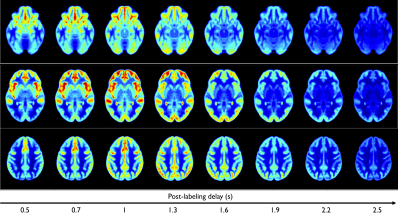
Finally, Fig5 shows the use of BATS to generate simulated ASL contrasts at various PLDs, showing consistent signal evolution based on transit-time and T1 distribution in the brain.
Conclusion
We have proposed an adult average perfusion template and ASL simulator based on high-resolution multi-parametric data. We have shown some early potential applications, such as direct registration of ASL data in case high-resolution anatomical scans aren’t available, but also the potential for versatile multi-contrast generation to cope with varying imaging parameters across sites, populations, enhancing possibilities for single-subject or large-scale neuroimaging analyses involving functional methods such as ASL. The BATS will be made publicly available shortly.Acknowledgements
No acknowledgement found.References
1. Good, C. D. et al. A voxel-based morphometric study of ageing in 465 normal adult human brains. Neuroimage 14, 21–36 (2001).
2. Collins, D. L., Neelin, P., Peters, T. M. & Evans, A. C. Automatic 3D Intersubject Registration of MR Volumetric Data in Standardized Talairach Space. Journal of Computer Assisted Tomography 18, 192–205 (1994).
3. Mazziotta, J. C., Toga, A. W., Evans, A., Fox, P. & Lancaster, J. A probabilistic atlas of the human brain: theory and rationale for its development. The International Consortium for Brain Mapping (ICBM). Neuroimage 2, 89–101 (1995).
4. Ashburner, J. & Friston, K. J. Voxel-based morphometry--the methods. Neuroimage 11, 805–21 (2000).
5. Mutsaerts, H. J. M. M. et al. ExploreASL: An image processing pipeline for multi-center ASL perfusion MRI studies. NeuroImage 219, 117031 (2020).
6. Avants, B. B. et al. The pediatric template of brain perfusion. Scientific Data 2, 1–17 (2015).
7. Taso, M., Munsch, F., Zhao, L. & Alsop, D. C. Regional and depth-dependence of cortical blood-flow assessed with high-resolution Arterial Spin Labeling (ASL). Journal of Cerebral Blood Flow & Metabolism (2020) doi:10.1177/0271678X20982382.
8. Avants, B. B. et al. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49, 2457–2466 (2010).
9. Avants, B. B. et al. A Reproducible Evaluation of ANTs Similarity Metric Performance in Brain Image Registration. Neuroimage 54, 2033–2044 (2011).
10. Tustison, N. J. & Avants, B. avants. Explicit B-spline regularization in diffeomorphic image registration. Front. Neuroinform. 7, (2013).
11. Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31, 968–980 (2006).
12. Buxton, R. B. et al. A general kinetic model for quantitative perfusion imaging with arterial spin labeling. Magn Reson Med 40, 383–396 (1998).
13. Alsop, D. C. & Detre, J. A. Reduced transit-time sensitivity in noninvasive magnetic resonance imaging of human cerebral blood flow. J. Cereb. Blood Flow Metab. 16, 1236–1249 (1996).
Figures