2663
Probing the Feasibility and Performance of Super-Resolution Head and Neck MRA Using Deep Machine Learning1Radiology, NorthShore University HealthSystem, Evanston, IL, United States, 2University of Chicago Pritzker School of Medicine, Chicago, IL, United States, 3Northwestern University Feinberg School of Medicine, Chicago, IL, United States
Synopsis
Deep machine learning approaches offer the potential for improved super-resolution (SR) reconstruction which could be useful in many clinical applications. Patients with suspected stroke often undergo MRI, which often includes magnetic resonance angiography (MRA) of the head and neck arteries with scan times of ≈10 to 15 minutes using standard nonenhanced methods. With the aim of shortening scan times, we evaluated the feasibility and performance of four deep neural network (DNN)-based SR reconstructions for restoring the image quality and spatial resolution of thin slab stack-of-stars quiescent interval slice-selective (QISS) head and neck MRA with degraded slice resolution.
Introduction
Vascular evaluation of the head and neck remains a key component in the diagnostic evaluation of patients presenting with suspected stroke.1 Recently, 3D thin-slab stack-of-stars quiescent interval slice-selective (3D tsSOS-QISS) MRA has been shown to provide high spatial resolution of the entire neck and Circle of Willis in ≈7 minutes with better image quality than time-of-flight (TOF) MRA.2 Nonetheless, further reduction of scan time would be desirable to improve patient comfort, reduce motion artifacts, and hasten diagnostic evaluation. Deep neural network (DNN)-based methods have recently found uses in numerous medical imaging applications, including for image reconstruction and restoration in accelerated MRI.3 We hypothesized that DNN-based SR reconstruction could be applied to 3D tsSOS-QISS head and neck MRA to allow the acquisition of a reduced number of thicker slices, thereby shortening scan time while preserving spatial resolution.Methods
General: This study was approved by our institutional review board and all participants (n=8, 7 healthy volunteers, 1 patient with carotid disease) provided written informed consent. Imaging was done using a prototype 3D tsSOS-QISS MRA sequence on a 3T MRI system (MAGNETOM Skyrafit, Siemens Healthineers, Erlangen) equipped with a 20-channel head and neck coil.3D tsSOS-QISS Protocol: Based on prior work2, prototype 3D tsSOS-QISS MRA of the head and neck was acquired with the following parameters: 0.86×0.86×1.30mm3 spatial resolution interpolated to 0.43×0.43×0.65mm3, axial coverage of 288.6mm, scan time 6min 39s, 300mm field of view, 352 acquisition matrix (704 reconstruction matrix after zero filling interpolation), fast low-angle shot readout with TR 9.9ms and TEs of 1.6ms, 3.7ms, and 5.7ms which were combined using a root mean square procedure2, QISS TR/QI of 1500/583ms. Acquired high-resolution (HR) volumes were considered as ground truth, while the 2-, 3-, 4-, 5-, and 6-fold lowered-resolution (LR) volumes were generated by Fourier transforming the HR data sets along the slice-direction, zeroing the most peripheral spatial frequencies along the slice direction, and inverse Fourier transformation back to the image domain.
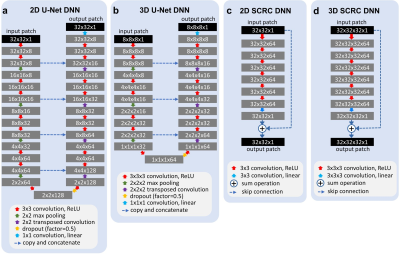
DNN Architectures: Four 2D patch/3D block-based deep neural network (DNN) models were tested: (1) 2D U-Net and (2) 3D U-Net4, and (3) 2D and (4) 3D networks consisting of serial convolutions and a residual connection (SCRC)5. These four DNNs, hereafter referred to as 2D U-Net, 3D U-Net, 2D SCRC, and 3D SCRC, respectively, are summarized in Figure 1.
Network Training: DNN training was done using leave-one-out cross-validation, an adaptive moment estimation optimizer, a mean squared error loss function, validation split of 20%, and early stopping based on validation loss. Typical training times for the 2D U-Net, 3D U-Net, 2D SCRC and 3D SCRC DNNs on a commodity graphics processing unit were ≈2 min (9 epochs), 9 min (7 epochs), 5 min (9 epochs) and 60 min (9 epochs), respectively.
Image Analysis: Images were analyzed quantitatively using Dice similarity coefficients (DSC) calculated for the intracranial and extracranial arteries, as well as through structural similarity index (SSIM), normalized root mean squared error (NRMSE), arterial sharpness, and arterial diameter measurements made in the bilateral M1 middle cerebral arteries. Forced choice scoring of randomized coronal maximum intensity projection (MIP) images (best DNN SR method versus LR input) was done by two experienced neuroradiologists.
Results
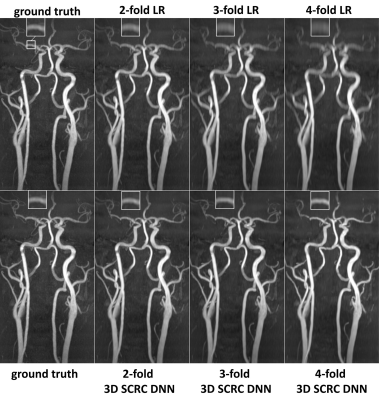
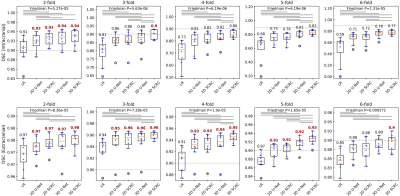
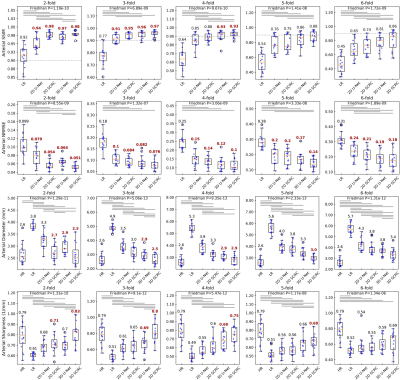
Figure 2 shows the image quality obtained with the 3D SCRC SR resolution technique with respect to the LR input volumes, and the target high-resolution output volume. With respect to the LR volume, 3D SCRC SR improved vessel conspicuity and sharpness in the slice-encoding (i.e. axial) direction, which mimicked that of the target volume for all tested resolution reduction factors.DSC measures of agreement in arterial anatomy portrayed in the SR-reconstructed and the ground truth volumes are shown in Figure 3. Arterial SSIM, NRMSE, diameter and sharpness metrics are summarized in Figure 4. All four quantitative metrics improved with application of the DNN SR techniques (Friedman, P<0.05), with arterial DSC, SSIM and sharpness values increasing, and with arterial NRMSE and diameter values decreasing with respect to those obtained from the LR volumes. Generally, 3D DNN outperformed 2D DNN SR reconstructions, while SCRC DNNs outperformed U-Net DNNs of the same dimensionality. The 3D SCRC DNN method provided the largest overall DSC values. The 3D DNNs provided median SSIM values of ≥0.9 and retained HR-levels of image sharpness for reduction factors of up to 4.
MR angiograms obtained with the best performing 3D SCRC SR DNN were consistently preferred by neuroradiologists over those obtained from the input LR volumes.
Discussion and Conclusion
We probed the capability of four DNN-based SR methods to improve spatial resolution in 3D tsSOS-QISS MRA configured with up to 6-fold degraded spatial resolution in the axial direction. We found that 2D and 3D DNN-based approaches restored image spatial resolution and appearance for resolution reduction factors up to 2 intracranially and 4 extracranially, according to multiple quantitative metrics including arterial DSC, SSIM, NRMSE, sharpness and diameter. In conclusion, DNN-based SR reconstruction can improve apparent spatial resolution in the axial direction and holds promise for substantially shortening the acquisition times of nonenhanced 3D tsSOS-QISS MRA.Acknowledgements
FUNDING SOURCE: NIH grant R01 EB027475References
1. Powers WJ, et al. Stroke. 2019
Dec;50(12):e344-e418. doi: 10.1161/STR.0000000000000211. Epub 2019 Oct 30.
2. Choy G,
et al. Radiology.
2018 Aug;288(2):318-328. doi: 10.1148/radiol.2018171820. Epub 2018 Jun 26.
3. Koktzoglou I,
et al. Magn Reson Med. 2020
Dec;84(6):3316-3324. doi: 10.1002/mrm.28339. Epub 2020 Jun 10.
4. Ronneberger O, Fischer P, Brox T. U-Net:
Convolutional Networks for Biomedical Image Segmentation. ArXiv150504597 Cs
2015.
5.
Chaudhari
AS, et al. Magn Reson Med. 2018 Nov;80(5):2139-2154. doi:
10.1002/mrm.27178. Epub 2018 Mar 26.
Figures