2661
Generalizability and Robustness of an Automated Deep Learning System for Cardiac MRI Plane Prescription1UC San Diego, La Jolla, CA, United States, 2GE Healthcare, Menlo Park, CA, United States, 3Mayo Clinic, Rochester, MN, United States, 4Mayo, Rochester, MN, United States
Synopsis
We show that an automated system of deep convolutional neural networks can effectively prescribe double-oblique imaging planes necessary for acquisition of cardiac MRI. To examine its clinical potential, we assess its component-wise performance by comparing simulated imaging planes predicted by DCNNs against ground truth imaging planes defined by a cardiac radiologist. Performance was assessed on 280 exams including 1252 image series obtained on ten 1.5T and 3T MRI scanners from three academic institutions. We further compare imaging planes acquired by technologists at the time of original acquisition against the same ground truth as an additional external reference.
Intro
Cardiac MRI is the reference standard for non-invasive assessment of cardiac morphology, function, and myocardial scar1. Broad accessibility of cardiac MRI is limited by the need for specially-trained physicians and technologists to plan dedicated double-oblique cardiac imaging planes2.Recently, deep convolutional neural networks (DCNNs) have shown promise for automating visual tasks in medical imaging, including localization of anatomic structures. More specifically, for cardiac MRI, previous groups showed the feasibility of utilizing DCNNs for the prescription of cardiac imaging planes3-5. However, one often cited limitation of machine learning algorithms, which may delay application in clinical practice, is uncertainty around generalizability, where an algorithm’s performance may vary outside the environment where it was trained6, 7. Moreover, as machine learning algorithms may occasionally fail, algorithm transparency is deemed essential to preserve confidence from its clinical end users.
With this in mind, we developed a multi-stage deep learning system to sequentially localize anatomic landmarks that define each of the cardiac imaging planes. This multi-stage approach provides visual feedback for each sequential step to preserve physician or technologist oversight. We then assessed the simulated component-wise performance of this deep learning system on cardiac MRIs obtained from multiple institutions to address concerns of algorithm generalizability.
Methods
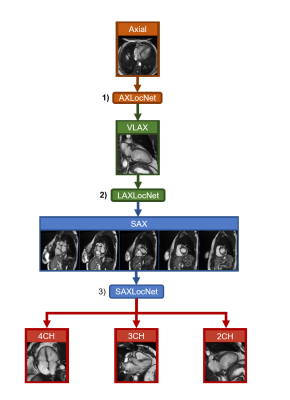
This was a HIPAA compliant, IRB approved retrospective study with waiver of informed consent. We developed a multi-stage system for prescribing cardiac imaging planes comprised of three DCNNs (AXLocNet, LAXLocNet, and SAXLocNet) to localize landmarks for prescription of long-axis, short-axis, and dedicated 4-, 3-, and 2-chamber views, respectively (Figure 1). Each DCNN was developed using U-Net-based heatmap regression. AXLocNet was a 3D U-Net trained with 4,647 image volumes, LAXLocNet was a 2D U-Net trained with 24,320 images, and SAXLocNet was a 3D U-Net trained with 11,109 image volumes.To assess generalizability, the algorithm was tested on an independent set of clinical exams from three tertiary academic medical centers across the United States, obtained from ten MRI scanners from a single vendor, including both 1.5T and 3T. A convenience sample of 280 examinations were included (129 from site A, 61 from site B, and 93 from site C) which included a total of 1,252 axial, long-axis and short-axis image series.
Ground truth imaging planes were defined on each image series by a board certified cardiac radiologist with over 10 years of experience in cardiac MRI. Imaging planes predicted by each DCNN component were compared against this ground truth by computing the difference in angulation. Similarly, imaging planes prescribed by the MRI technologist at the time of the examination were also compared against this ground truth to provide an additional point of reference. Wilcoxon-paired tests were used to assess for statistical significance.
Results
Long-axis imaging planes predicted by AXLocNet had a mean angulation error of 9.52±13.09°, which was greater than technologist angulation error 5.06±6.26° (p<0.001, n=219), with some site-to-site variation (Figure 2). Short-axis imaging planes predicted by LAXLocNet had a mean angulation error of 3.15±6.07°, which was lower than technologist angulation error 3.75±3.73°(p<0.001, n=280) (Figure 3). All three imaging planes predicted by SAXLocNet were comparable or had lower mean angulation errors than the technologist angulation error. For the 4-chamber view, SAXLocNet angulation error was 5.61±4.74°, while technologist error was 7.19±6.25° (p<0.001, n=201). For the 3-chamber view, SAXLocNet angulation error was 6.73±5.77°, while technologist error was 7.08±5.92° (p=0.637, n=178). For the 2-chamber view, SAXLocNet angulation error was 5.18±4.49°, while technologist error was 8.46±6.74° (p<0.001, n=172) (Figure 4).Discussion
Component-wise performance of the automated deep learning system was comparable to or exceeded performance of cardiac MRI technologists for the majority of cardiac imaging planes. Performance of the first component, AXLocNet, was an exception and showed greater mean angulation error. This may be partially explained by greater heterogeneity of cardiac morphologies and cardiac orientations within the thorax on axial imaging, compared to more standardized presentation of VLAX and SAX images. Future work may be directed at enhancing the performance of AXLocNet with more diverse training data or DCNN training strategies.While prior approaches for planning cardiac MRI imaging planes have focused primarily on planning all the imaging planes from a single axial acquisition, the multi-component approach proposed here provides opportunity for real-time supervision and adjustment, which may facilitate ready integration into existing clinical systems4, 5. This may facilitate greater accessibility and reliability of cardiac MRI by reducing the need for highly-specialized personnel for image acquisition, and democratize access to this important clinical modality. Future work may be directed to assessing improvements in image quality and potential reductions in scan time during prospective clinical acquisitions.
Conclusions
This study illustrates the generalizability and robustness of a multi-stage DCNN system to automate the planning of cardiac MRI imaging planes based on images from a broad cross-section of patients on MRI scanners from multiple institutions.Acknowledgements
No acknowledgement found.References
1. La Gerche A, Claessen G, Van de Bruaene A, et al. . Cardiac MRI: a new gold standard for ventricular volume quantification during high-intensity exercise . Circ Cardiovasc Imaging 2013; 6 (2): 329 – 338
2. Ferguson M, Otto R. . Cardiac MRI Prescription Planes . MedEdPORTAL 2014. ; 10 : 9906 . Accessed May 28, 2018
3. Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016 May;30:108-119. doi: 10.1016/j.media.2016.01.005. Epub 2016 Feb 6. PMID: 26917105.
4. Addy O, Jiang W, Overall W, Santos J, Hu B. . Autonomous CMR: Prescription to Ejection Fraction in Less Than 3 Minutes. Pacific Grove, Calif: International Society for Magnetic Resonance in Medicine Machine Learning Workshop , 2018. . https://cds.ismrm.org/protected/Machine18/program/videos/30950/
5. Oksuz et al., "Automatic left ventricular outflow tract classification for accurate cardiac MR planning," 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, 2018, pp. 462-465, doi: 10.1109/ISBI.2018.8363616.
6. Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, et al. (2018) Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLOS Medicine 15(11): e1002683. https://doi.org/10.1371/journal.pmed.1002683
7. Anushri Parakh, Hyunkwang Lee, Jeong Hyun Lee, Brian H. Eisner, Dushyant V. Sahani , Synho Do (2019) Urinary Stone Detection on CT Images Using Deep Convolutional Neural Networks: Evaluation of Model Performance and Generalization Radiology AI 1(4)
Figures
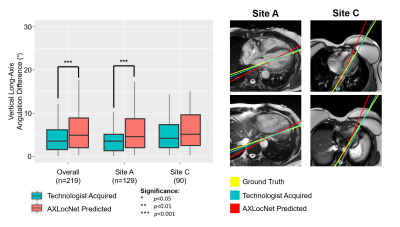
Left: Comparison of plane angulation differences from vertical long-axis planes acquired by an MRI technologist (teal) or AXLocNet (coral).
Right: Exemplar axial images displaying radiologist ground truth (yellow), technologist acquired (teal), and AXLocNet predicted (red) vertical long-axis planes at the level of the mitral valve (top) and apex (bottom). Ground truth and AXLocNet predicted localizations are shown as dots yellow and red, respectively. Due to too few images from site B (n=8), we excluded these from the subset analysis.
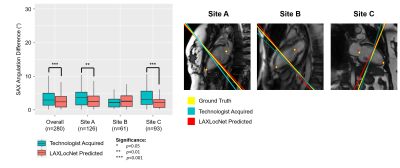
Left: Comparison of plane angulation differences from short-axis planes acquired by an MRI technologist (teal) or LAXLocNet (coral).
Right: Exemplar vertical long-axis images displaying radiologist ground truth (yellow), technologist acquired (teal), and LAXLocNet predicted (red) short-axis planes. Ground truth and LAXLocNet predicted localizations are shown as dots yellow and red, respectively.
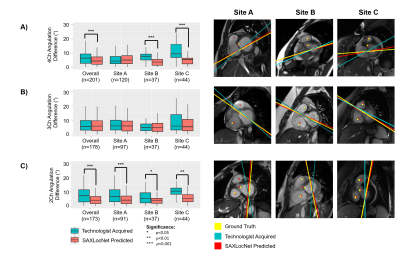
Left: Comparison of plane angulation differences from A) 4-chamber, B) 3-chamber, or C) 2-chamber planes acquired by an MRI technologist (teal) or SAXLocNet (coral).
Right: Exemplar vertical long-axis images displaying radiologist ground truth (yellow), technologist acquired (teal), and SAXLocNet predicted (red) A) 4-chamber, B) 3-chamber, or C) 2-chamber planes. Ground truth and SAXLocNet predicted localizations are shown as dots yellow and red, respectively.