2656
Cardiac MRI feature tracking by deep learning from DENSE data1Biomedical Engineering, University of Virginia, Charlottesville, VA, United States, 2Radiology, University of Virginia, Charlottesville, VA, United States
Synopsis
As cine DENSE provides myocardial contours and intramyocardial displacement data, we investigated the use of DENSE to train deep networks to predict intramyocardial motion from contour motion. FlowNet2, an optical-flow convolutional neural network, was used as a comparator/reference, and as the starting point for a DENSE-trained network (DT-FlowNet2). Further, we added a correction network with convolution along time, resulting in a through-time-corrected DENSE-trained network (TC-DT-FlowNet2). TC-DT-FlowNet2 outperformed other methods, providing accurate intramyocardial displacements from myocardial contours. DENSE-based learning of intramyocardial displacements from contours holds promise as a new method for computing strain from the contours of standard cine MRI.
Introduction
MRI myocardial strain imaging is used diagnostically and prognostically for many types of heart disease. There are multiple methods for performing strain MRI, each with its own advantages and disadvantages. Feature tracking (FT) is the most widely used and convenient method, as it applies post-processing algorithms to standard cine images to assess strain; however, it is less accurate, reproducible and prognostic than strain-dedicated acquisitions like displacement encoding with stimulated echoes (DENSE)1-3. FT directly tracks myocardial contours, but does not track intramyocardial features, as the myocardium on standard cine MRI generally produces a very uniform signal and doesn’t present intramyocardial features suitable for tracking. Instead, the intramyocardial motion is (imperfectly) estimated using optical-flow based methods applied to the time series of endocardial and epicardial contours4. In contrast, DENSE directly measures intramyocardial tissue displacement, leading to its high accuracy and better clinical performance; however, this method requires the acquisition of additional images dedicated to strain assessment, which adds time to imaging studies. As DENSE images provide both myocardial contours and intramyocardial displacement data, we investigated the use of DENSE data to train deep networks to predict intramyocardial motion from contour motion. This deep learning (DL) approach may provide more accurate intramyocardial displacements (the precursor to strain) than optical-flow-based methods when applied to myocardial contour data.Methods
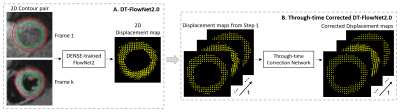
Our study design made use of a very successful optical-flow convolutional neural network (CNN), called FlowNet25, which is widely used for frame-to-frame motion tracking in video imagery. Because conventional FT algorithms are not published, we used FlowNet2 as a comparator/reference for other methods. Also, we built upon FlowNet2 by using it as our starting point and fine-tuning it using DENSE training data. To train DENSE-trained FlowNet2 (DT-FlowNet2), the input was the endocardial and epicardial contours in two image frames, and the output was the frame-to-frame displacement field (Fig. 1A). The first of the two frames was always the end-diastolic image, as end diastole is always the reference time for DENSE.We also exploited the temporal dimension of multiphase cine MRI by adding a correction network with convolution along time, resulting in a through-time-corrected DENSE-trained network (TC-DT-FlowNet2). The through-time convolution used a 3D kernel with 2D spatial and 1D temporal dimensions. The input of the correction network was a stack of sequential displacement fields from DT-FlowNet2 with size . The factor of 2 accounts for displacements in two directions, and are the sizes of DENSE images and represents the number of temporal frames. We treated the 2D displacements as two channels in the input, and the output was the corrected displacement fields (Fig. 1B). The correction network has 6 convolution layers with number of filters being 2, 32, 96, 96, 96, 2, respectively. All convolution kernels are of size (3,3,3). Datasets were normalized by the maximum value in each subject, and the final results were scaled back by multiplying by the scalar. Because the output size of DT-FlowNet2 and the input size of the 3D correction network were not matched, they were trained separately.
Our training data is from 108 subjects, with each subject including three short-axis slices of cine DENSE images encoded for two-dimensional in-plane displacement. Half of the datasets are from healthy volunteers, and the other half includes patients with myocardial infarction, heart failure, or dilated cardiomyopathy. After removing slices of poor quality, we had 286 total cine DENSE slices. For DT-FlowNet2, DENSE data from 155 slices (6674 image pairs) were used for training and 131 slices (6061 image pairs) for testing. For TC-DT-FlowNet2, 105 slices were used for training, and 26 for testing, fully using the 131 test slices output from DT-FlowNet2.
Results
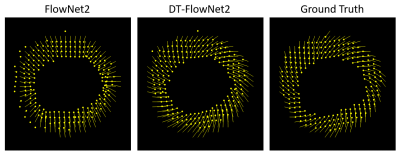
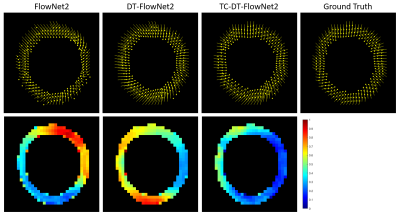
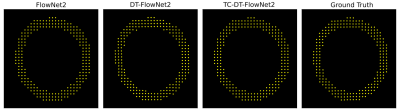
DT-FlowNet2 showed better agreement with the ground truth than unmodified FlowNet2. An example is shown in Fig. 2, illustrating that DT-FlowNet2 correctly depicts intramyocardial twisting while FlowNet2 incorrectly depicted radially-oriented displacement. End-point-error (EPE), which represents the difference of two displacement vectors, was used for error calculation and comparisons. Overall, DT-FlowNet2 showed a lower EPE than FlowNet2 (0.69±0.18 vs. 0.80±0.25, p<0.01). The through-time correction further improved performance, as TC-DT-FlowNet2 provided a lower EPE than DT-FlowNet2 (0.51±0.16 vs. 0.60±0.18, p<0.01) on 26 test slices. Comparisons including the TC-DT-FlowNet2 are shown in Fig. 3, demonstrating further improved accuracy, including lower time-averaged error (Fig. 3, bottom row) compared to ground truth. Example myocardial displacement movies comparing the various methods are shown in Fig. 4, which clearly show the advantage of the through-time correction.Conclusion
The use of 2D spatial DENSE training data along with a through-time correction provided low displacement error compared to ground truth, and showed the ability to depict detailed intramyocardial motion patterns such as twist, which are otherwise challenging for optical-flow based methods applied to contour data. DL of intramyocardial displacement from contour motion using DENSE datasets holds promise as a potentially improved method for computing strain from the contours of standard cine MRI. Future work will evaluate TC-DT-FlowNet2 for analysis of contours derived from standard cine MRI.Acknowledgements
This work was supported by R01HL147104.References
- Lin K, Meng L, Collins JD, Chowdhary V, Markl M, Carr JC. Reproducibility of cine displacement encoding with stimulated echoes (DENSE) in human subjects. Magnetic resonance imaging. 2017;35:148-53.
- Bilchick KC, Auger DA, Abdishektaei M, Mathew R, Sohn M-W, Cai X, et al. CMR DENSE and the Seattle heart failure model inform survival and arrhythmia risk after CRT. JACC: Cardiovascular Imaging. 2020;13(4):924-36.
- Mangion K, Carrick D, Carberry J, Mahrous A, McComb C, Oldroyd KG, et al. Circumferential strain predicts major adverse cardiovascular events following an acute ST-segment–elevation myocardial infarction. Radiology. 2019;290(2):329-37.
- Schuster A, Hor KN, Kowallick JT, Beerbaum P, Kutty S. Cardiovascular magnetic resonance myocardial feature tracking: concepts and clinical applications. Circulation: Cardiovascular Imaging. 2016;9(4):e004077.
- Ilg E, Mayer N, Saikia T, Keuper M, Dosovitskiy A, Brox T. Flownet 2.0: Evolution of optical flow estimation with deep networks. InProceedings of the IEEE conference on computer vision and pattern recognition 2017 (pp. 2462-2470).
Figures