2640
Unsupervised Tag Removal in Cardiac Tagged MRI using Robust Variational Autoencoder1Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Division of Cardiology, Children's Hospital Los Angeles, Los Angeles, CA, United States
Synopsis
Post processing of cardiac tagged MRI has always been challenging because of poor SNR and image artifact. Inspired by unsupervised anomalies detection using variational autoencoder (VAE), we treat tags as anomalies and employ robust variational autoencoder (RVAE), using \(\beta\)-ELBO cost, which is more robust to outliers, to replace the log-likelihood optimization of a prototypical VAE, to generate tag-free results from cardiac tagged images.
Introduction
Tagged Magnetic Resonance Imaging (tMRI), placing tags such as stripes or grids on tissue using radiofrequency saturation pulses, measures the deformation of tissue, typically in the heart1. However, post processing of cardiac tMRI has always been challenging because of poor SNR and image artifact2. To recover high quality cardiac anatomic images from tMRI, several tag removal methods have been proposed. Band-pass3 and band-stop4 filters are easy to implement, but results vary with filter design and parameter setting, and images are often overly smoothed or blurry due to the loss of high frequency information. A deep learning approach5, modeling tag removal as image inpainting problem by treating tags as corrupted region and employing a two-stage generative adversarial network to recover the images6, can generate high quality tag-free results. However, the network uses supervised learning strategy and requires extracting tagged region first as mask, which could cause weak generalization due to the different tagging patterns. Recently, variational autoencoder (VAE)7, as an unsupervised method, has been increasingly applied in image synthesis, denoising, and anomalies detection. In this paper, we treat tags as anomalies and employ robust variational autoencoder (RVAE)8, using \(\beta\)-ELBO cost, which is more robust to outliers, to replace the log-likelihood optimization of a prototypical VAE, to generate tag-free results from cardiac tagged images.Methods
Robust Variational Autoencoder: Let \(x^{(i)}\in R^{D}\) be an observed sample of input data \(X\), where \(i\in\left\{ 1,2,...,N\right\}\) , \(D\) is the dimensionality of the feature space, and \(N\) is the sample size. \(z^{(i)}\) is an observed sample from latent space \(Z\), where \(j\in\left\{ 1,2,...,S\right\}\) . Let \(q_{\theta}(Z\mid X)\) and \(p_{\phi}(X\mid Z)\) denote encoder and decoder distribution, respectively, where \(\theta\) and \(\phi\) are the network parameters. The loss function of VAE can be represented as equation (1), where the first term can be interpreted as reconstructed loss, and the second term is KL divergence9 regularizer. $$L_{VAE}(\theta,\phi;x^{(i)})=-E_{q_{\theta}(Z \mid x^{(i)})}[log(p_{\phi}(x^{(i)} \mid Z))]-D_{KL}(q_{\theta}(Z \mid x^{(i)})\parallel p_{\phi}(Z))\;\;\;(1)$$We can replace the reconstructed loss with \(\beta\)-cross entropy10 shown in equation (2), and assume Gaussian noise statistics. The loss function of RVAE can be expressed as equation (3), where \(\beta\) is a hyperparameter between 0 to 1. The selection of \(\beta\) depends outlier proportion in the dataset. $$H_\beta^{(i)}(\hat{p}(X),p_{\theta}(X\mid z^{(j)}))=-\frac{\beta+1}{\beta}\int_{}^{}\delta(X-x^{(i)})(N(\hat{x}^{(j)},\sigma)^{\beta}-1)dX+\int_{}^{} N(\hat{x}^{(j)},\sigma)^{\beta+1}dX\;\;\; (2)$$ $$L_{\beta}(\theta,\phi;x^{(i)})=\frac{\beta+1}{\beta}(\frac{1}{(2\pi\sigma^{2})^{\beta D/2}}exp(-\frac{\beta}{2\sigma^2}\sum_{d=1}^D\parallel\hat{x}_d^{(j)}-x_d^{(j)}\parallel^2)-1)-D_{KL}(q_{\theta}(Z \mid x^{(i)})\parallel p_{\phi}(Z))\;\;\;(3)$$
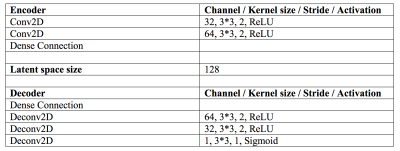
Network Architecture and Parameters: The architecture details are shown in Table 1. To speed up the feature extraction and avoid encoding redundant image details such as noise and artifact, the encoder uses a light design only containing two convolution layers. We set the size of latent space to 128 and use three deconvolution layers in the decoder to guarantee the image quality of outputs. According to our observation, the tag region usually occupies 10 to 20 percentage of the tissue. Therefore, we set \(\beta\) = 0.001 in our experiments.
Data Description and Training Procedure: Secondary use of the clinical data was approved by the IRB. 30 subjects contained both tagged images and reference images with 20 frames in each cardiac cycle. Four chamber tagged spoiled gradient echo images and steady state free precession reference images were acquired on a 1.5T General Electric CVi system. To control for differences in contrast, we used reference images as ground truth, and linearly registered the tags onto reference images to generate simulated tagged images so that the tag removal performance can be quantitively evaluated by the comparison between recovered images and ground truth. The dataset was split into the training and test set with a ratio of 4:1. The network was implemented in TensorFlow 2.0 and trained frame by frame in 2D images. We choose the ADAM optimizer with a learning rate of 0.001 and run the experiments on two NVIDIA Tesla P100 GPUs.
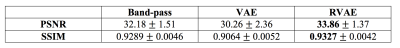
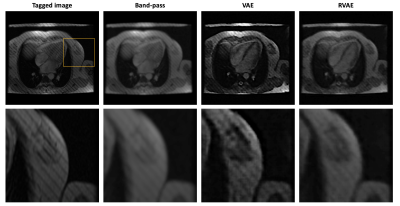
Evaluation: To quantitively evaluate the tag removal performance, the experiment was run on the simulated tagged data, and the results were compared with the reference images. We used a filtering based method, band-pass3, as baseline, and compare RVAE with the baseline and the prototypical VAE. General image quality metrics, peak signal-to-noise ratio (PSNR) and structural similarity index (SSIM), are used to evaluate the quality of reconstructed images. To explore the feasibility of RVAE in practical tag removal task, we also run prediction directly on real tagged images, and visually compared the image results.
Results and Discussion
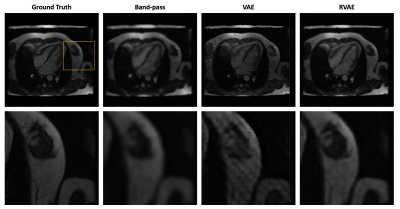
Fig.1 and Fig.2 show the tag removal results on simulation experiment and real tagged images. Compared to VAE and RVAE, band-pass method severely blurs the images due to the loss of high frequency information. The results of VAE are generally sharper than other two. However, the tags are preserved because the log-likelihood term in the cost function is sensitive to the outliers. Therefore, VAE might not be feasible for tag removal. Due to the robustness of β-divergence10, RVAE can generate tag-free images although the results are slightly smoother than the ones of VAE. The corresponding quantitative results shown in table 2 indicate that VAE has the worst performance in tag removal, because the preserved tags cause larger reconstructed error and structural dissimilarity, while RVAE achieves both higher PSNR and SSIM than other methods. The outperformance of RVAE proves its effectiveness of retrieving tagging corrupted images.Acknowledgements
This work was supported in part by the National Heart Lung and the Blood Institute under Grant 1U01HL117718-01 and Grant 1RO1HL136484-A1, and in part by the National Center for Research Resources under Grant UL1 TR001855-02. Computation for the work described in this abstract was supported by the University of Southern California’s Center for High-Performance Computing (hpcc.usc.edu).References
1. Zerhouni EA, Parish DM, Rogers WJ, Yang A, Shapiro EP. Human heart: Tagging with MR imaging--a method for noninvasive assessment of myocardial motion. Radiology 1988;169(1):59-63.
2. Makram AW, Rushdi MA, Khalifa AM, El-Wakad MT. Tag removal in cardiac tagged MRI images using coupled dictionary learning. IEEE EMBC. 2015; 7921-7924.
3. Osman NF, Kerwin WS, McVeigh ER, Prince JL. Cardiac motion tracking using CINE harmonic phase (HARP) magnetic resonance imaging. Magnetic Resonance in Medicine. 1999;42(6):1048-60.
4. Qian Z, Huang R, Metaxas D, Axel L. A novel tag removal technique for tagged cardiac MRI and its applications. IEEE ISBI. 2007; 364-367.
5. Chai Y, Xu B, Zhang K, Lepore N, Wood J. MRI Restoration Using Edge-Guided Adversarial Learning. IEEE Access. 2020; PP. 1-1. 10.1109/ACCESS.2020.2992204.
6. Xu B, Chai Y, Wood J. Tag Removal in Cardiac Tagged MRI using Edge-guided Adversarial Learning. ISMRM 2020. 2232
7. Kingma DP and Welling M. An introduction to variational autoencoders. arXiv preprint arXiv:1906.02691.
8. Akrami H, Joshi AA, Li J, Aydore S, Leahy RM. Robust variational autoencoder. arXiv preprint arXiv:1905.09961.
9. Joyce JM. Kullback-Leibler Divergence. In: Lovric M. (eds) International Encyclopedia of Statistical Science. 2011. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-04898-2_327
10. Basu A, Harris I, Hjort N, Jones M. Robust and Efficient Estimation by Minimising a Density Power Divergence. Biometrika, 85(3), 549-559. 1998. Retrieved December 16, 2020, from http://www.jstor.org/stable/2337385
Figures