2638
Comparison of Traditional fSNAP and 3D FuseUnet Based fSNAP1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China
Synopsis
By adapting 3D FuseUnet, CNN fSNAP showed better performance in lumen and IPH depiction compared with traditional fSNAP. The results suggest that deep learning can help fast SNAP scans produce high quality images, which could have great clinical utility.
Introduction
Carotid atherosclerosis is a major cause of ischemic stroke. Carotid stenosis and intraplaque hemorrhage (IPH) are the most import morphology and composition characteristics in determining the vulnerability of carotid atherosclerosis. The SNAP sequence imaging has been proved to have high sensitivity in detection of IPH (1,2). However the imaging speed of SNAP is slow due to the additional acquired Ref-TFE for background suppression compared with traditional MPRAGE sequence. To accelerate SNAP imaging, fSNAP with low-resolution reference scan to estimate the background phase was proposed and validated. Though, fSNAP (3) can have a similar result with SNAP and provide 37.5% shorter scan time than SNAP, the image quality of fSNAP can would be hampered when a high downsampling factor is applied to the Ref-TFE, resulting in ringing artifact, inaccurate background phase and so on. Inspired by the outstanding performance of CNN in medical image reconstruction/processing, we aim to use CNN to further increase SNAP Ref-TFE downsampling without losing image quality. In this study, we trained a CNN network that achieved a better image quality than fSNAP.Methods
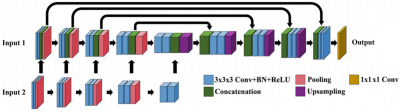
1) Dataset: We retrospectively recruited 133 patients with recent ischemic symptom. All patients were scanned using a 3T whole body scanner (Philips, Achieva) with SNAP sequence and the real image, imaginary image were generated for both IR-TFE and Ref-TFE of SNAP. 107 cases without IPH from the cohort were random chosen for training and the rest were used for testing. In order to balance the data of IPH patients and normal people, and to verify the model's ability to distinguish between IPH and lumen on the test set, the IPH patients were randomly divided into training+validating and test date set in a balance manner. And the IPH data in training dataset was separately augmented. 2)Data preprocessing: Fourier transform was applied to the fully acquired SNAP Ref-TFE. The Ref-TFE was downsampling with 16×16 factor in the kspace keeping the kspace center region to simulate a low-resolution Ref-TFE. A fSNAP corrected real (CR) and corrected imaginary (CI) image set could be generated with fully acquired IR-TFE and low-resolution Ref-TFE using phase sensitive reconstruction. 3)Network architecture: We follow the 3D FuseUnet framework as shown in Figure 1, which consists of a Unet and an extra Feature extraction network, and perform feature fusion on different feature scales so that the network makes full use of image domain information. The real and imaginary parts of IR-TFE and fSNAP are used as input 1 and input 2 respectively and the loss for the network is MSE. In addition, in order to improve the spatial perception of the network, all operations are three-dimensional, with a 192*192*32 patch size.RMSE was calculated for fSNAP and FuseUnet (CNN) fSNAP respectively. And the RMSE was compared with paired t-test using GraphPad Prisim.
Results
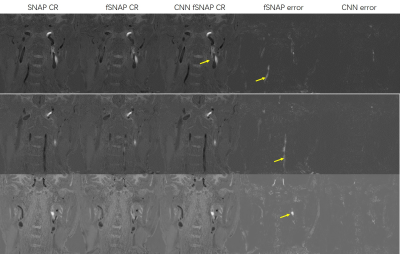
We demonstrate the effectiveness of the proposed method with test set images. As shown in Figure 2, with an extremely high downsampling rate (16×16), the performance of CNN fSNAP is better than traditional fSNAP. The CNN fSNAP performance is significant better thant traditional fSNAP (RMSE 66.87 vs. 202.58, p<0.001). Visually, the result from CNN fSNAP, which is simliar to the original image is obviously better than fSNAP.Discussion
By adapting 3D FuseUnet, CNN fSNAP showed better performance in lumen and IPH depiction compared with traditional fSNAP. The results suggest that deep learning can help fast SNAP scans produce high quality images, which could have great clinical utility. Further study would include the fine tuning the current network with prospectively downsampled in-vivo data.Acknowledgements
References
1. Wang J, Bornert P, Zhao H, et al. Simultaneous Noncontrast Angiography and intraPlaque (SNAP) Imaging for Carotid Atherosclerotic Disese Evaluation. Magn Reson Med. 2013;69:337-345.
2. Li D, Qiao H, Han Y,et al. Histological validation of simultaneous non-contrast angiography and intraplaque hemorrhage imaging (SNAP) for characterizing carotid intraplaque hemorrhage. Eur Radiol. 2020.
3. Chen S, Ning J, Zhao X, et al.Fast Simultaneous Noncontrast Angiography and Intraplaque Hemorrhage (fSNAP) Sequence for Carotid Artery Imaging. Magn Reson Med. 2017;77:753–758.