2636
The Comparison of denoising methods for cardiac diffusion tensor imaging
Xi Xu1, Yuxin Yang1, Yuanyuan Liu1, Dong Liang1, Hairong Zheng1, and Yanjie Zhu1
1Shenzhen Institute of Advanced Technology, ShenZhen, China
1Shenzhen Institute of Advanced Technology, ShenZhen, China
Synopsis
We evaluate three different image denoising methods in cardiac diffusion tensor imaging (CDTI) regarding image quality and accuracy of parameter estimates with simulation and ex-vivo experiments. The local principal component analysis (LPCA) performs the best in improving image quality both in simulated and ex-vivo data, and the uncertainty of parameter estimations is reduced by all three algorithms in the ex-vivo experiment.
Introduction
Cardiac diffusion tensor imaging (CDTI) is a powerful tool that realizes the non-invasive detection of myocardial microstructure.1 Several indices extracted from CDTI have shown the potential to identify microstructural abnormalities, including fractional anisotropy (FA), mean diffusivity (MD), helix angle (HA), and absolute secondary vector angulation (E2A).2-3 However, the inherently low signal-to-noise ratio (SNR) of CDTI causes inaccurate estimation of these parameters. Several denoising methods have been proposed for brain DTI, based on the assumption of Rician distribution or the multi-directionality of DW images, such as adaptive nonlocal mean (ANLM), 4 local principal component analysis (LPCA), 5 Marchenko–Pastur principle component analysis (MPPCA). 6 The above methods can also be applied for CDTI, but their performance is unknown. In this study, we evaluate the performance of the above three algorithms in CDTI in simulation and ex-vivo experiments by the SNR of the image and the accuracy of the four parameters (FA, MD, HA, and E2A) after denoising.Methods
In the simulation, we use the dataset downloaded from the IEEE dataport, including CDTI images and diffusion tensors (D). 7 The simulated reference image (S0) is generated by setting the values in the myocardium to 1000 and the background to 0. Then diffusion images () are simulated according to the Eq. (1):$$S_{DWI}=S0\ast e^{-b\ast g^{T}\ast D\ast g}$$ (1)
where g is the gradient vectors, b is the b-value = 350 s/mm2. Then white Gaussian noise is added to the simulated CDTI images with SNR = 5, 10, 15, 20, and 25. The SNRs of the denoised images using ANLM, LPCA, and MPPCA are calculated. CDTI parameters are computed from the noise-free and denoised images, respectively, using a home-made software. The root-mean-square error (RMSE) between them evaluates the parameter estimation accuracy.
In the ex-vivo experiment, the CDTI data of 37 slices of the failing heart replaced from four patients are acquired on a 3T scanner (Ingenia, Philips, The Netherlands). The study is approved by the Medical Research Ethics Committee (No.KY20200390101) of Guangdong Provincial People's Hospital. Imaging parameters are turbo spin-echo DTI sequence with 16 diffusion encoding directions, b-value=800 s/mm2, repetition = 8. The average images of 8 repetitions with LPCA denoising and the corresponding indices are used as the gold standard. The average images with repetition = 1, 2, 4, 6, 8 are used to imitate images with different SNR levels. The three methods are then applied on average images. The SNRs of denoised images are calculated as the mean signal intensity in the myocardium divided by the standard deviation in the background. CDTI parameter maps are computed from denoised images and the gold standard, then RMSE between them indicates accuracy.
Results
In the simulation, ANLM and LPCA produce clear images, while MPPCA can hardly reduce noise (Fig. 1). Evident inhomogeneity can be seen in the noisy map, then the parameters get more accurate after denoising from the LPCA and ANLM (Fig 2). In the ex-vivo experiment, the improvement of LPCA is obvious, and the SNR is higher than the other two methods. MPPCA performs worst as the residual noise exists obviously (Fig. 3). As the SNR increases, the RMSE of all parameters and algorithms shows a downward trend (Fig. 4, 5). Combined with visual judgment, LPCA can achieve apparent homogeneity, and consistent with the previous study, 5 the most significant reduction in uncertainty of parameter estimates is obtained using the LPCA method than another method.Discussion and conclusion
The three post-processing methods improve image quality and accuracy in parameter estimates in the ex-vivo experiment. The ANLM performs well in low SNR but seems to over smooth some image details. The MPPCA removes only thermal noise, resulting in apparent noise residue. The LPCA reduces the noise and preserves image contrast, and produces accurate parameters that reflect the tissue's characteristics. We suggest using LPCA to improve image quality and parameter estimations accuracy in CDTI imaging.Acknowledgements
This work is supported in part by the National Natural Science Foundation of China under grant nos. 61771463,81971611, National Key R&D Program of China nos. 2020YFA0712202, 2017YFC0108802 , the Innovation and Technology Commission of the government of Hong Kong SAR under grant no. MRP/001/18X, and the Chinese Academy of Sciences program under grant no. 2020GZL006..References
- Nielles‐Vallespin, Sonia, et al. "Cardiac diffusion: technique and practical applications." Journal of Magnetic Resonance Imaging 52.2 (2020): 348-368.
- Khalique, Zohya, and Dudley Pennell. "Diffusion tensor cardiovascular magnetic resonance." Postgraduate Medical Journal 95.1126 (2019): 433-438.
- Ferreira, Pedro F., et al. "In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy." Journal of Cardiovascular Magnetic Resonance 16.1 (2014): 87.
- Manjón, José V., et al. "Adaptive nonlocal means denoising of MR images with spatially varying noise levels." Journal of Magnetic Resonance Imaging 31.1 (2010): 192-203.
- Manjón, José V., et al. "Diffusion weighted image denoising using overcomplete local PCA." PloS one 8.9 (2013): e73021.
- Veraart, Jelle, Els Fieremans, and Dmitry S. Novikov. "Diffusion MRI noise mapping using random matrix theory." Magnetic resonance in medicine 76.5 (2016): 1582-1593.
- Verzhbinsky, Ilya A., et al. "Estimating Aggregate Cardiomyocyte Strain Using In~ Vivo Diffusion and Displacement Encoded MRI." IEEE Transactions on Medical Imaging 39.3 (2019): 656-667.
Figures
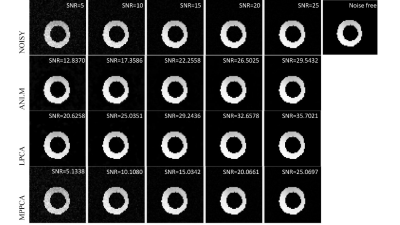
Fig.1 Simulated
DWI image corresponding to a noise-free simulation randomly selected, the noisy
image of SNR = 5~25 after adding Gaussian white noise, and the results after
denoising

Fig.2 Parameter
estimations of helix angle (HA) from the simulated, noisy and denoised (ANLM,
LPCA, MPPCA) images.
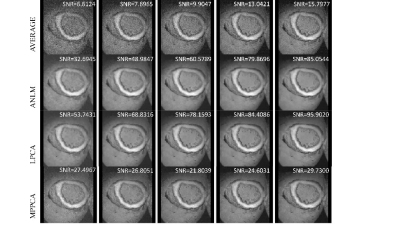
Fig.3 Representative
noisy and denoised images from ex vivo data. The image on the 1st row is averaged 1, 2, 4, 6, 8,
respectively. The 2nd row ANLM perform very well but seems over
smoothed, then 3rd-row LPCA produced clear image with details
preserved, the last row MPPCA is slightly better than the average of 8-time
with noise residue.
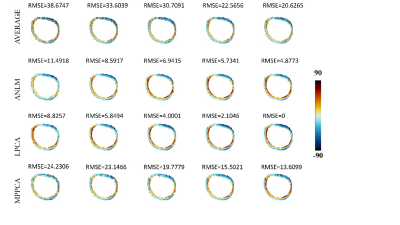
Fig.4 Parameter estimations
of helix angle (HA) from ex-vivo data.
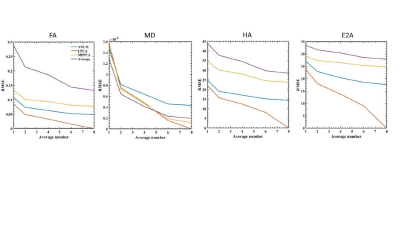
Fig.5 RMSEs
of the CDTI parameter estimates of the simulation as a function of the SNR