2634
Automatic segmentation of middle cerebral artery plaque based on deep learning1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 2College of Software, Xinjiang University, Urumqi, China, 3Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 4CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 5Department of radiology, Beijing Chao-Yang hospital, Capital medical university, beijing, China
Synopsis
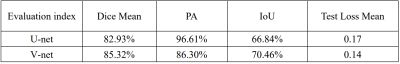
At present, deep learning has gradually been applied to the field of plaque segmentation. However, the existing work is mainly used for the processing of 2D images. In this study, we trained a 3D network model to automatically segment the middle cerebral artery plaques based on 3D images and compared the accuracy with 2D network model. Magnetic resonance vessel wall imaging (MR-VWI) data from 102 patients were used for training. The results showed that all quantitative accuracy indicators of V-net were higher than U-net, and experiments showed that V-net was more stable.
Introduction
Cerebral arterial atherosclerotic plaque rupture is a major cause of ischemic stroke [1-2]. Magnetic resonance vessel wall imaging (MR-VWI) is currently the only non-invasive imaging method for directly imaging and characterizing the cerebral arterial atherosclerotic plaques [3]. The traditional manual quantification is time-consuming, laborious, and heavily dependent on the doctor's experience. At present, deep learning has gradually been applied to the field of plaque segmentation [4-5]. Deep learning can greatly improve the accuracy of plaque recognition and classification, reduce the workload of doctors, and improve diagnosis efficiency. However, the existing work is mainly used for the processing of 2D images. In this study, we trained a 3D network model to segment the plaques based on 3D images and compared the accuracy with 2D network model.Materials and Methods
Data Acquisition: A total of 102 patients with plaques identified on middle cerebral arteries (MCA) were recruited to the study. All participants underwent MR-VWI using a 3D SPACE sequence on a 3.0 Tesla whole-body MR system (MAGNETOM Prism; Siemens Healthcare, Erlangen). Two radiologists reviewed in consensus and manually segmented the MCA plaques on the MR-VWI images of the 102 patients using an open-source software ITK-SNAP (version 3.8.0, www.itk-snap.org). And regions of interest (ROIs) were drawn automatically in the center of the segmentation result area. The box was resized to 64x64 as input for deep learning networks.Training models: Two deep learning methods: 2D U-net and 3D V-net were used for the automatic segmentation of the plaques [6-7]. They have similar institutions, which were a fully connected convolutional residual network and consists of convolution and max-pooling layers at the descending part (the left component of model), and convolution and up-sampling layers at the sending part (the right component of model). The difference between them is that U-net analyzes 2D data and V-net analyzes 3D data, which can combine the relationships between different layers. The segmentation performance was evaluated using the Dice, pixel accuracy (PA), and Intersection-over-Union (IoU).
Results
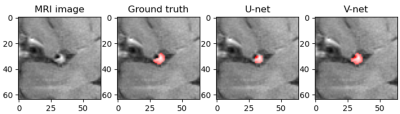
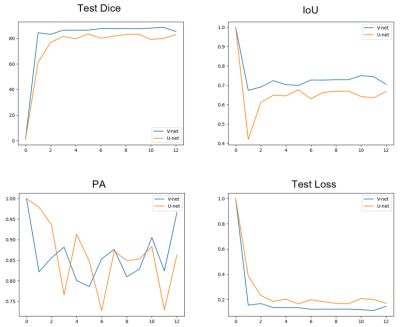
Representative images of the segmentation results of the two deep learning models (U-net and V-net) was shown in Figure 1. Table 1 summarized the three quantitative indicators (mean dice, PA and IoU) of the model to reflect the accuracy of the results. Machine learning achieved stable performance as shown by mean Dice, the mean Dice of U-net reached 82.93%, and the mean Dice of V-net reached 85.32%. The curve of loss decreasing with training is shown in Figure 2. The correlation between the prediction output of the model and the training epoch of each group is shown in Figure 3. Both models can effectively complete the segmentation of atherosclerosis. In addition, all parameters of V-net are higher than U-net, and experiments show that V-net is more stable.Discussion
The study verifies the effectiveness of using neural networks to segment cerebral artery plaques. Machine learning on atherosclerotic plaques of MRI can accurately segmentation the edge of plaques. This study has the following limitations. The first was its relatively small sample size; especially in the test cohort, a larger data set is needed to evaluate this prediction model in the future. Secondly, the data input to the model is cropped rather than the entire MRI image. As more data sets become available, these limitations will be resolved. Deep learning can automatically analyze and generate diagnostic recommendations. It can assist doctors in diagnosis, improve the accuracy of the diagnosis of brain plaque, and reduce the workload of doctors. and the results may be used to develop a fully automatic diagnostic tool that can be implemented for clinical use.Acknowledgements
The study was partially support by National Natural Science Foundation of China (81830056),Capital Health Development Research Project (2020-2-2037), Natural Science Foundation of Guangdong Province (2018A030313204), and Shenzhen Basic Research Program (JCYJ20180302145700745 and KCXFZ202002011010360).References
[1] G. Pasterkamp et al. Atherosclerotic plaque rupture: an overview. Paper presented at: Journal of Clinical & Basic Cardiology, 2000. 3 (2): p.705-713
[2] Holmstedt, Christine A et al. Atherosclerotic intracranial arterial stenosis: risk factors, diagnosis, and treatment. Paper presented at: The Lancet. Neurology vol. 12,11 (2013): 1106-14.
[3] Slijkhuis, W et al. A historical perspective towards a non-invasive treatment for patients with atherosclerosis. Paper presented at: Netherlands heart journal : monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation vol. 17,4 (2009): 140-4.
[4] Maier, Andreas et al. A gentle introduction to deep learning in medical image processing. Paper presented at: Zeitschrift fur medizinische Physik vol. 29,2 (2019): 86-101.
[5] Esteva, Andre et al. Dermatologist-level classification of skin cancer with deep neural networks. Paper presented at: Nature vol. 542,7639 (2017): 115-118.
[6] Ronneberger et al. U-net: Convolutional networks for biomedical image segmentation. Paper presented at: International Conference on Medical image computing and computer-assisted intervention 2015.
[7] Lei, Yang et al. Ultrasound prostate segmentation based on multidirectional deeply supervised V-Net. Paper presented at: Medical physics vol. 46,7 (2019): 3194-3206.
Figures