2632
SVD-Based Multi-Channel-Receive-Coil Combination for 13C Metabolic Imaging1GE Healthcare, Munich, Germany, 2Department of Radiology, University of Cambridge, Cambridge, United Kingdom, 3MR Research Centre, University of Aarhus, Aarhus, Denmark
Synopsis
Multi-channel receive coils can improve coverage for metabolic imaging with hyperpolarised 13C compounds. Combining different receive channels in an SNR-optimal way is challenging due to the difficulties in determining sensitivity maps. The main aim of this work was to implement and optimise a Singular-Value-Decomposition (SVD) based sensitivity map extraction from metabolic images with a single spectral point per metabolite and to investigate its performance in SNR-limited metabolic imaging experiments.
Introduction
Multi-channel receive coils can improve coverage for metabolic imaging with hyperpolarised 13C compounds. Especially when imaging larger regions of interest, such as the human abdomen, these coils gained popularity. The downside of multi-channel receive coils is a more involved reconstruction: SNR-optimal coil combination requires receive coil sensitivity maps, which are not readily available for 13C because of its low natural abundance signal. Therefore, root-mean-squares coil combination is popular due to its ease of implementation and robustness. More SNR-optimal methods extract sensitivity maps from the acquired metabolic data itself, and use those for phasing, weighting and combining the different coil elements [1,2]. These methods were applied to MRSI data with full spectroscopic sampling. The goal of the present work was to implement and optimise the SVD-based sensitivity map extraction from metabolic images with a single spectral point per metabolite and investigate its performance in heavily SNR-limited experiments.Methods
Experimental:Kidney, heart and breast data were acquired with IDEAL-spiral Chemical-Shift Imaging (CSI), where a single-shot spiral trajectory is delayed in different excitations to encode a few spectral points [3]. Brain data was acquired using spectral-spatial excitation combined with a single-shot spiral readout [4]. Experiments were performed at two different centres on whole-body 3T scanners (MR750, GE Healthcare, Waukesha, WI, USA) equipped with various multi-channel 13C receive coils (Rapid Biomedical, Rimpar, Germany). [1-13C]pyruvate was polarised using a SPINlab Polariser (GE Healthcare, Waukesha, WI, USA) and injected in humans and pigs. The acquired data were reconstructed automatically on the scanner using spectral inversion (only for IDEAL), spatial apodisation, zero-filling and gridding reconstruction, yielding 5 metabolic images for pyruvate, lactate, alanine, bi-carbonate and pyruvate-hydrate.
Coil combination:
The reconstructed data was fed into the coil-combination algorithm consisting of the following steps: (1) averaging data along the time dimension; (2) noise decorrelation of data; (3) SVD-based calculation of sensitivity maps; (4) background phase removal of sensitivity maps; (5) polynomial interpolation for smoothing of sensitivity maps; (6) normalisation of sensitivity maps to a root-mean-squares value of one along the coil dimension; (7) multiplication of data with sensitivity maps to weight and phase individual coil maps spatially; (8) summation of data (both original and time-averaged) along the coil dimension. For comparison, coils were also combined via root-mean-square (RMS) calculation along the coil dimension.
Time-averaging the data acquired over multiple time steps (step 1) is possible, because the phase does not change over time, hence reducing matrix size for the SVD calculation. The noise decorrelation (step 2) uses the lowest 10% of the (not time averaged) signal in spatial domain as noise, assuming that the object is not covering the full field of view. The SVD calculation (step 3) uses the complex data (with matrix size #time-steps*#metabolites by #coils) looping over all spatial voxel. The complex coil sensitivities are given directly by the first right singular vector. The resulting phase of these complex coil sensitivities has phase jumps depending on the object/coil arrangement, which are removed through the background phase removal (step 4): a signal-weighted phase average is calculated over all coils; this spatially-varying pseudo-phase is then subtracted from the sensitivity maps. Polynomial interpolation of the complex coil sensitivities (step 5) is applied to smoothen the sensitivity maps [5]. Normalisation (step 6) is required, because the “body coil” image is not available, and sensitivity maps must not induce additional, data-dependent signal. In the final step (7), the original data is corrected by the coil sensitivity maps and summed over the coil dimension.
The signal-to-noise ratio was determined by calculating the mean of the highest 3% of signal over the standard deviation of the lowest 10% of signal from each respective metabolite image.
Results and Discussion
Coil combination of metabolic images using the SVD-based method removes the Rician noise bias of the RMS method and improves SNR, particularly when the metabolic images are noisy (Figs. 1,3,4,5). Applying noise decorrelation and polynomial smoothing (subsequently called “SVD+”) to the data further improves SNR. In the human 13C kidney data (Fig. 1) lactate SNR was improved from 52 (RMS), 55 (bare SVD) to 62 (SVD+) for lactate and from 123, 132 to 142 for pyruvate. SNR values for the other organs are listed in the respective captions (Fig. 3-5). Sensitivity maps extracted from the data look reasonable, especially after polynomial smoothing (Fig. 2).Conclusion
SVD-based coil combination was shown to be robust and improved SNR in multiple datasets and could be included in the automatic reconstruction on the MRI scanner.Acknowledgements
No acknowledgement found.References
[1] Receive array magnetic resonance spectroscopy: Whitened singular value decomposition (WSVD) gives optimal Bayesian solution. Rodgers CT, Robson MD. Magn Reson Med. 2010 Apr;63(4):881-91. doi: 10.1002/mrm.22230.
[2] Coil combination methods for multi-channel hyperpolarized 13C imaging data from human studies. Zhu Z, Zhu X, Ohliger MA, Tang S, Cao P, Carvajal L, Autry AW, Li Y, Kurhanewicz J, Chang S, Aggarwal R, Munster P, Xu D, Larson PEZ, Vigneron DB, Gordon JW. J Magn Reson. 2019 Apr;301:73-79. doi: 10.1016/j.jmr.2019.01.015.
[3] IDEAL spiral CSI for dynamic metabolic MR imaging of hyperpolarized [1-13C]pyruvate. Wiesinger F, Weidl E, Menzel MI, Janich MA, Khegai O, Glaser SJ, Haase A, Schwaiger M, Schulte RF. Magn Reson Med. 2012 Jul;68(1):8-16. doi: 10.1002/mrm.23212.
[4] Saturation-recovery metabolic-exchange rate imaging with hyperpolarized [1-13C] pyruvate using spectral-spatial excitation. Schulte RF, Sperl JI, Weidl E, Menzel MI, Janich MA, Khegai O, Durst M, Ardenkjaer-Larsen JH, Glaser SJ, Haase A, Schwaiger M, Wiesinger F. Magn Reson Med. 2013 May;69(5):1209-16. doi: 10.1002/mrm.24353.
[5] SENSE: sensitivity encoding for fast MRI. Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. Magn Reson Med. 1999 Nov;42(5):952-62
Figures
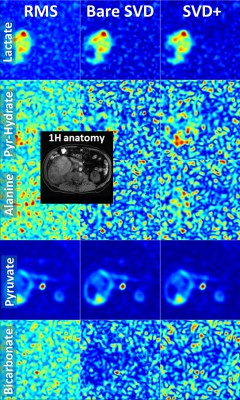
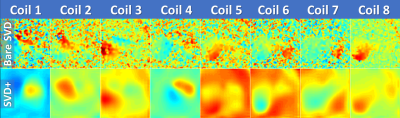
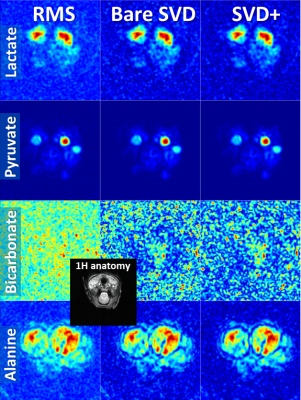
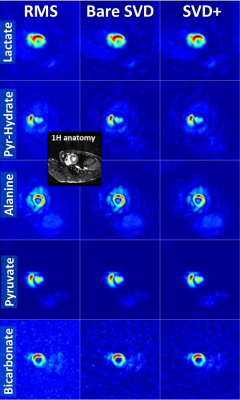
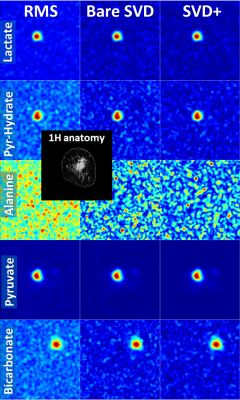
Fig. 5: Comparison of different coil combinations in a breast cancer patient acquired with an 8-channel receive coil (average of time steps 3 to 10; slice 2 out of 2). The field-of-view of the 13C images covers the left breast only. The spatially shifted bicarbonate signal stems from the external bicarbonate reference phantom placed next to the breast. SNR increased from RMS, to bare SVD to SVD+ for lactate: 31, 40, 48; pyruvate-hydrate: 28, 32, 36; alanine: 24, 24, 27;pyruvate: 164, 196, 252; bicarbonate: 29, 35, 40.