2598
Accelerated Chemical Exchange Saturation Transfer Acquisition by Joint K-space and Image-space Parallel Imaging (KIPI)1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 2MR Collaboration, Siemens Healthcare Ltd., Shanghai, China
Synopsis
The clinical use of chemical exchange saturation transfer (CEST) imaging is limited by its relatively long scan time, because it typically acquires multiple saturation image frames. Here, a novel auto-calibrated reconstruction method by joint k-space and image-space parallel imaging (KIPI) is proposed for faster CEST acquisition. By under-sampling CEST image frames with variable acceleration factors, KIPI allows an acceleration factor of up to 8-fold for acquiring source images, yielding a net speed-up of 6-fold in scan time, and produces image quality close to that of the ground truth.
Introduction
Chemical exchange saturation transfer (CEST) is an emerging MRI technique that amplifies the detectability of certain low-concentration biomolecules through their interaction with the abundant water pool.1,2 However, its routine clinical use is limited by the relatively long scan time since CEST MRI acquires multiple image frames.3,4 Recently, a variably-accelerated SENSE (vSENSE) method was proposed to speed up the CEST acquisition.5,6 Here, we propose a novel auto-calibrated reconstruction method by joint k-space and image-space parallel imaging (KIPI) as a further development of the vSENSE approach.Theory
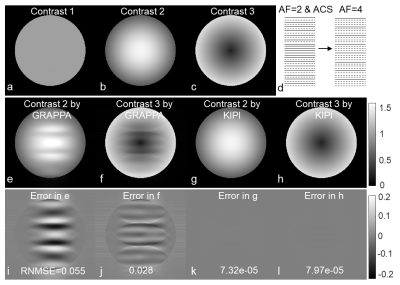
As an auto-calibrated k-space reconstruction method, GRAPPA7 estimates the missing k-space point by a linear combination of the acquired data in the neighborhood from all channels using the following equation,$$\begin{equation}y=Aw\tag{1} \end{equation}$$where $$$A$$$ represents the source matrix organized from the auto-calibration signal (ACS) data, $$$y$$$ denotes the target matrix, and $$$w$$$ represents the weights to be fitted. GRAPPA generates accurate images robustly when the acceleration factor (AF) is low.8 However, the GRAPPA weights estimated can be affected by the underlying image contrast. Figure 1 shows images of different contrasts simulated according to Biot-Savart law9 with eight receive channels. It is clear that the GRAPPA weights calculated from one contrast yield significant errors when being applied to the other contrasts (Figs. 1e-1f and 1i-1j).SENSE10 poses the reconstruction as a linear inverse problem in image space and theoretically yields the optimal solution. However, the accuracy of SENSE reconstruction depends on the accuracy of the sensitivity maps used, which are prone to errors in practice.5,6 If the coil image $$$m_{i}$$$ of channel $$$i$$$ ($$$1\leq i\leq N$$$) is perfectly known, the sensitivity map $$$SE_{i}$$$ can be obtained by dividing the coil-combined image $$$\rho$$$ into the image from each channel.$$\begin{equation}SE_{i}=m_{i}/\rho\tag{2}\end{equation}$$
Thus, for retroactive SENSE reconstruction with AF=2 (without loss of generality), there is $$\begin{equation}\begin{split}\begin{aligned}s_{N*1}&=m_{N*1}^{1}+m_{N*1}^{2}\\&=\rho^{1}SE_{N*1}^{1}+\rho^{2}SE_{N*1}^{2}\\&=\left[\begin{array}{ll}SE_{N*1}^{1}&SE_{N*1}^{2}\end{array}\right]\left[\begin{array}{l}\rho^{1}\\\rho^{2}\end{array}\right]\end{aligned}\end{split} \tag{3}\end{equation}$$ where $$$s_{N*1}$$$ represents the folded coil image vector, and the superscript represents the spatial position. The least squares solution to Eq. [3] is$$\begin{equation}\begin{split}\begin{aligned}\left[\begin{array}{c}\hat{\rho}^{1}\\\widehat{\rho}^{2}\end{array}\right]&=\left(SEE^{H}SEE\right)^{-1}SEE^{H}s_{N*1}\\&=\underbrace{\left(SEE^{H}SEE\right)^{-1}SEE^{H}SEE}_{I}\left[\begin{array}{l}\rho^{1}\\\rho^{2}\end{array}\right]=\left[\begin{array}{l}\rho^{1}\\\rho^{2}\end{array}\right]\end{aligned}\end{split} \tag{4}\end{equation}$$ where $$$SEE=\left[\begin{array}{ll}SE_{N*1}^{1}&\left.SE_{N*1}^{2}\right]\end{array}\right.$$$ and $$$I$$$ is the identity matrix.
However, the coil image $$$m_{i}$$$ is unknown for SENSE imaging with AF > 1 in practice. The KIPI method calculates $$$m_{i}$$$ with GRAPPA reconstruction from a low-AF calibration frame. Then, the sensitivity maps can be calculated with Eq. [2], and be applied to the other frames with high-AF SENSE reconstruction. Notably, the derivation above (Eqs. [2-4]) illustrates that the SENSE solution using the sensitivity maps derived from the calibration frame itself is identical to its GRAPPA solution.
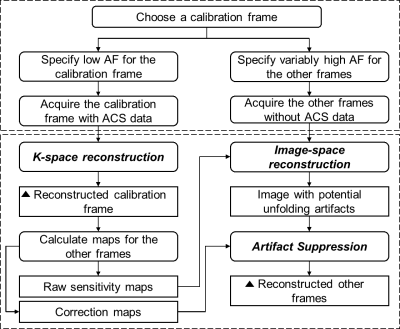
Overall, KIPI turns the original vSENSE approach into an auto-calibrated parallel imaging method by incorporating both GRAPPA and SENSE reconstruction. During data sampling, KIPI chooses an image frame as the calibration frame with low AF with the ACS data, and undersamples the other image frames using variably high AF without ACS data. During reconstruction, KIPI inherits the robustness of auto-calibrated GRAPPA imaging, and avoids the drawback of its susceptibility to underlying contrasts by calculating sensitivity maps. Furthermore, KIPI can adopt the artifact suppression algorithm used in vSENSE.5,6 The KIPI approach is summarized in Figure 2.
Methods
Human experiments were conducted on a 3T Siemens Prisma MRI system using a 64-channel-receive head coil. A SPACE-CEST sequence11 was run using the following readout parameters: FOV=212×212×201mm3, resolution=2.8×2.8×2.8mm3, turbo factor=140, and GRAPPA factor=2×2 with ACS acquired. Seven CEST saturation offsets for APT-weighted (APTw) imaging were executed, including (S0), ±3, ±3.5, and ±4-ppm. And a dual-echo 3D GRE sequence was used for B0 field mapping.For retrospective reconstruction, the +3.5-ppm frame was selected as the calibration frame with AF=2×2 to generate the sensitivity maps and correction maps, and the remaining 6 frames had AF=2×4 (in the phase-encoding and partition-encoding directions, respectively). Note that only the calibration frame retained ACS data.
Results
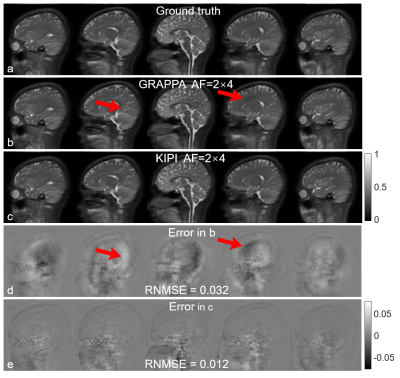
Figures 1g-h show KIPI allowed a calibration frame with a different image contrast to be used, generating reconstruction errors substantially smaller than those from GRAPPA (Figs. 1k-l vs. 1i-j). On the contrary, the accuracy of GRAPPA was comprised when reusing the kernel weights across different image contrasts (Figs. 1e-f).The conventional GRAPPA (AF=2×2) scan (Fig. 3a) was considered ground truth here. KIPI (Fig. 3c) generated source images that better agreed with the ground-truth results than those reconstructed from GRAPPA (Fig. 3b) using the same data (AF=2×4) with RNMSE of 0.012 versus 0.032, respectively. Besides, unfolding artifacts were evident in the GRAPPA images (Figs. 3b, d; red arrows).
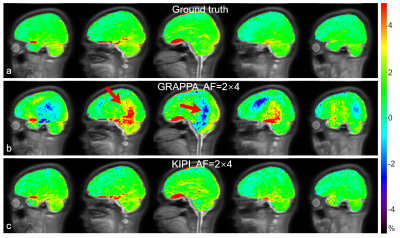
Figure 4 displays the APTw images calculated from the source images of Fig. 3 after B0 correction and image alignment. Substantial artifacts can be seen in the APTw images reconstructed by GRAPPA (Fig. 4b) while the KIPI method generated APTw maps (Fig. 4c) that were consistent with the ground truth (Fig. 4a).
Conclusion
KIPI is a novel auto-calibrated parallel imaging method that incorporates the advantages of k-space and image-space reconstruction methods. KIPI allowed an acceleration factor of up to 8-fold for acquiring CEST source images, yielding a net speed-up of 6-fold in scan time, and essentially without compromising the image quality. KIPI should facilitate the translation of CEST imaging in clinical routines.Acknowledgements
NSFC grant number: 81971605, 61801421.References
1.Ward KM, Aletras AH, Balaban RS. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn. Reson. 2000;143(1):79-87.
2.van Zijl PCM, Yadav NN. Chemical Exchange Saturation Transfer (CEST): What is in a Name and What Isn't? Magn Reson Med 2011;65(4):927-948.
3.Zhou JY, Lal B, Wilson DA, et al. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med 2003;50(6):1120-1126.
4.Zhou J, Blakeley JO, Hua J, et al. Practical data acquisition method for human brain tumor amide proton transfer (APT) imaging. Magn Reson Med 2008;60(4):842-849.
5.Zhang Y, Heo H-Y, Lee D-H, et al. Chemical Exchange Saturation Transfer (CEST) Imaging With Fast Variably-accelerated Sensitivity Encoding (vSENSE). Magn Reson Med 2017;77(6):2225-2238.
6.Zhang Y, Heo H-Y, Jiang S, et al. Fast 3D chemical exchange saturation transfer imaging with variably-accelerated sensitivity encoding (vSENSE). Magn Reson Med 2019;82(6):2046-2061.
7.Griswold MA, Jakob PM, Heidemann RM, et al. Generalized Autocalibrating Partially Parallel Acquisitions (GRAPPA). Magn Reson Med 2002;47(6):1202-1210.
8.Blaimer M, Breuer F, Mueller M, et al. SMASH, SENSE, PILS, GRAPPA: how to choose the optimal method. Top Magn Reson Imaging 2004;15(4):223-236.
9.Guerquin-Kern M, Lejeune L, Pruessmann KP, et al. Realistic Analytical Phantoms for Parallel Magnetic Resonance Imaging. IEEE Trans Med Imaging 2012;31(3):626-636.
10.Pruessmann KP, Weiger M, Scheidegger MB, et al. SENSE: Sensitivity encoding for fast MRI. Magn Reson Med 1999;42(5):952-962.
11.Zhang Y, Yong X, Liu R, et al. Whole-brain chemical exchange saturation transfer imaging with optimized turbo spin echo readout. Magn Reson Med 2020;84(3):1161-1172.
Figures